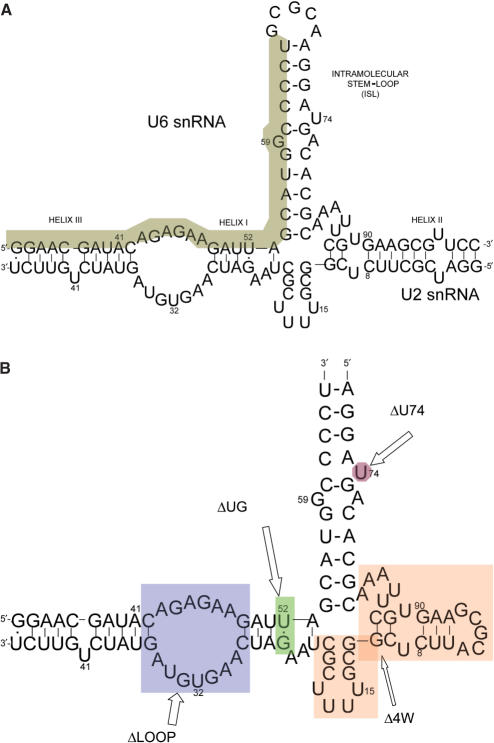
Figure 2.
Sequences of RNA oligomers used in this work. (A) Proposed complex formation between the central domains of human U2 and U6 snRNA. Previously established helices I, II, III and U6 intra-molecular stem loop (ISL) are labeled. The invariant top strand 32-mer area is shaded. (B) The simplified construct and its mutations used in the FRET study. The sequence represents the simplified original construct (called WT), which includes an invariant 32-nt top strand and a 69-nt bottom strand. Fluorescent dyes are attached at the 3′ or 5′ end of the 32-nt strand. The four mutation areas are shaded. All mutations were created by variations of segments of the bottom strand. ΔLOOP eliminates the ACAGAGA loop by mutating the bottom sequence to result in a complementary stem. ΔUG mutates the U–G base pair to U–A. Δ4W eliminates the four-way junction by replacing the shaded area with three adenosines. ΔU74 deletes the shaded uridine from the U6 ISL sequence.