Abstract
HIV infects macrophages and microglia in the central nervous system (CNS), which express lower levels of CD4 than CD4+ T cells in peripheral blood. To investigate mechanisms of HIV neurotropism, full-length env genes were cloned from autopsy brain and lymphoid tissues from 4 AIDS patients with HIV-associated dementia (HAD). Characterization of 55 functional Env clones demonstrated that Envs with reduced dependence on CD4 for fusion and viral entry are more frequent in brain compared to lymphoid tissue. Envs that mediated efficient entry into macrophages were frequent in brain, but were also present in lymphoid tissue. For most Envs, entry into macrophages correlated with overall fusion activity at all levels of CD4 and CCR5. gp160 nucleotide sequences were compartmentalized in brain versus lymphoid tissue within each patient. Proline at position 308 in the V3 loop of gp120 was associated with brain compartmentalization in 3 patients, but mutagenesis studies suggested that P308 alone does not contribute to reduced CD4 dependence or macrophage-tropism. These results suggest that HIV adaptation to replicate in the CNS selects for Envs with reduced CD4 dependence and increased fusion activity. Macrophage-tropic Envs are frequent in brain but are also present in lymphoid tissues of AIDS patients with HAD, and entry into macrophages in the CNS and other tissues is dependent on the ability to use low receptor levels and overall efficiency of fusion.
Keywords: Human Immunodeficiency Virus Type 1 (HIV), envelope, brain, lymph node, CD4, neurotropism
Introduction
Human immunodeficiency virus type 1 (HIV) infects the central nervous system (CNS) in the early stages of disease (An et al., 1999; Davis et al., 1992) and causes HIV-associated dementia (HAD) or milder forms of neurocognitive impairment in 10–20% of AIDS patients (Gonzalez-Scarano and Martin-Garcia, 2005; Kaul et al., 2001; Kaul et al., 2005; McArthur et al., 2003; Sacktor et al., 2002). Highly active anti-retroviral therapy (HAART) has reduced the incidence of HAD (d'Arminio Monforte et al., 2004; Dore et al., 1999; Sacktor et al., 2001; Sacktor et al., 2002), but the prevalence of minor cognitive and motor disorders (MCMD) has been increasing as HIV-infected individuals survive for a longer time on anti-retroviral therapies (Dore et al., 2003; McArthur et al., 2003; Neuenburg et al., 2002; Valcour et al., 2004). Most current antiviral drugs have relatively poor CNS penetration. Thus, the CNS is an important reservoir for long-term viral persistence and neurocognitive impairment continues to be a significant complication of HIV disease.
HIV enters the CNS via trafficking of infected monocytes and lymphocytes across the blood-brain barrier (reviewed in (Dunfee et al., 2006a; Gonzalez-Scarano and Martin-Garcia, 2005). The main target cells for HIV infection in the CNS are macrophages and microglia. HIV entry is initiated by the viral envelope glycoprotein gp120 binding to CD4. Structural rearrangements induced by CD4 binding allow subsequent interaction with a chemokine coreceptor, usually CCR5 or CXCR4, leading to further conformational changes and fusion of viral and target cell membranes (Deng et al., 1996; Dragic et al., 1996; Oberlin et al., 1996; Trkola et al., 1996; Wu et al., 1996). Coreceptor usage is a major determinant of cellular tropism. CCR5 using (R5) viruses infect CD4+ T cells and macrophages, and CXCR4 using (X4) viruses infect CD4+ T cells and T cell lines. However, CCR5 usage alone does not predict macrophage tropism (M-tropism) (Cheng-Mayer et al., 1997; Cunningham et al., 2000; Dittmar et al., 1997; Gorry et al., 2001; Hung et al., 1999). A subset of R5 viruses do not productively infect macrophages (Gorry et al., 2001; Li et al., 1999). Furthermore, some M-tropic HIV strains use CXCR4 for entry in macrophages and microglia (Ancuta et al., 2001; Gorry et al., 2001; Koning et al., 2001; Naif et al., 2002; Simmons et al., 1998; Singh et al., 2001; Yi et al., 1999; Yi et al., 1998). Receptor density also determines the susceptibility of primary cells and cell lines to HIV infection (Fear et al., 1998; Kuhmann et al., 2000; Platt et al., 1998; Rana et al., 1997; Reynes et al., 2001; Tuttle et al., 1998). Thus, while CCR5 is the main coreceptor for HIV entry in macrophages, viral determinants independent of those that influence coreceptor usage also influence macrophage tropism.
Macrophages and microglia express lower levels of CD4 than CD4+ T cells in peripheral blood (Di Marzio et al., 1998; Dick et al., 1997; Graziani-Bowering and Filion, 2000; Jordan et al., 1991; Kazazi et al., 1989; Lewin et al., 1996; Martin-Garcia et al., 2002; Peudenier et al., 1991a; Peudenier et al., 1991b; Tuttle et al., 1998; Wang et al., 2002; Williams et al., 1992). Previous studies identified a few brain-derived HIV viruses from HAD patients that replicate efficiently in macrophages and microglia and have reduced dependence on CD4 and/or CCR5 levels for entry (Gorry et al., 2001; Gorry et al., 2002; Martin-Garcia et al., 2006; Peters et al., 2004). Reduced CD4 or CCR5 dependence may therefore contribute to HIV neurotropism and neurovirulence. However, whether HIV Envs that have reduced dependence on CD4 and CCR5 levels are more frequent in brain compared to other tissues remains unclear.
Genetic evolution of HIV within the brain is distinct from that in lymphoid tissues and other organs (Chang et al., 1998; Donaldson et al., 1994; Gartner et al., 1997; Gorry et al., 2001; Hughes et al., 1997; Korber et al., 1994; Shapshak et al., 1999; van't Wout et al., 1998; Wong et al., 1997). Genetic compartmentalization of viral variants in the brain suggests that adaptive evolution may occur in the CNS in response to unique constraints of the brain microenvironment, such as different target cell populations and immune selection pressures. Strain et al reported several residues in the C2-V3 region of Env associated with compartmentalization in the cerebrospinal fluid (CSF) (Strain et al., 2005). However, other studies failed to identify sequence differences associated with brain compartmentalization (Ohagen et al., 2003; Reddy et al., 1996), and Env sequences that enhance HIV neurotropism have not been identified.
To investigate genetic and functional characteristics of Envs in brain versus lymphoid tissues, full-length env genes were amplified from autopsy brain and lymphoid tissues from 4 AIDS patients with HAD. Envs with reduced dependence on CD4 levels were more frequent in brain, but were also present in lymphoid tissue. The capacity of Envs from both brain and lymphoid tissues to mediate entry into macrophages correlated with overall fusion activity at all levels of CD4 and CCR5. Proline at position 308 in the V3 region of gp120 was associated with brain compartmentalization in 3 patients, but mutagenesis studies suggested that P308 alone did not contribute to the reduced CD4 dependence or macrophage-tropism of brain-derived Envs. Thus, HIV adaptation to replicate in the CNS microenvironment selects for Envs with reduced CD4 dependence and increased fusion activity. Macrophage-tropic Envs are frequent in brain but are also present in lymphoid tissues of HAD patients, and entry into macrophages is influenced by the capacity to use low receptor levels and overall efficiency of fusion.
Results
HIV envelopes with reduced dependence on CD4 levels are more frequent in brain compared to lymphoid tissues
To determine whether HIV Envs with reduced dependence on CD4 or CCR5 levels are more frequent in brain compared to lymphoid tissues, full-length HIV env genes were cloned directly from autopsy brain (frontal lobe or basal ganglia) and lymphoid (lymph node or spleen) tissues from 4 AIDS patients with HAD (MACS2, MACS3, UK1, and UK7) (Table 1). Eight to 40 Env clones from each tissue sample were screened for functional activity in single-round infection assays by pseudotyping onto NL43 env-GFP and detecting infection of CCR5/CXCR4 positive Hela/CD4 cells (clone JC53). By this approach, 10 tissue samples from 4 AIDS patients yielded 55 functional Env clones (n=35 brain and 20 lymphoid). All 55 Env clones used CCR5 to enter cells, 2 weakly used CCR3, and none used CXCR4 (Table 1). None of the Env clones mediated CD4-independent infection in Cf2 cells expressing CCR5. This pattern of coreceptor usage is consistent with the R5 phenotype of primary viruses previously isolated from these tissue samples ((Gorry et al., 2001) and P. Gorry and D. Gabuzda, unpublished data).
TABLE 1.
Characteristics of patients and envelope clones
| Patient | Risk Factora | CD4 countb | Antiretroviral therapy | HIV encephalitis | Giant cells | Autopsy tissuec | Viral loadd | No. of clones | Coreceptor usagee |
|---|---|---|---|---|---|---|---|---|---|
| MACS2 | MH | 52 | AZT | Moderate | + | FL | 1200 | 8 | R5 (R3) |
| LN | 8700 | 3 | R5 | ||||||
| SP | 2400 | 3 | R5 | ||||||
| MACS3 | MH | 95 | None | Moderate | + | FL | 660 | 8 | R5 |
| LN | 100800 | 6 | R5 | ||||||
| UK1 | IVDU | 87 | ddc (1mo) | Moderate | ++ | FL | 51500 | 7 | R5 |
| BG | 44800 | 4 | R5 | ||||||
| SP | 1400 | 6 | R5 | ||||||
| UK7 | IVDU | 90 | AZT | Severe | ++ | FL | 27800 | 8 | R5 |
| LN | 300 | 2 | R5 |
MH, male homosexual; IVDU, intravenous drug user.
Cells per μl.
FL, frontal lobe; BG, basal ganglia; LN, lymph node; SP, spleen.
HIV Gag DNA copies per 106cells. Viral load for MACS2FL, UK1FL and UK1BG is an average of the viral load for 2 samples.
Coreceptor usage was determined by infection of Cf2 cells expressing CD4 and either CCR5, CXCR4 or CCR3. Parenthesis indicates weak coreceptor usage.
To investigate the frequency of Envs in brain versus lymphoid tissue that can use low levels of CD4 or CCR5 to mediate fusion, we performed cell-to-cell fusion assays with Cf2 cells expressing low (Mean Fluorescent Intensity (MFI) 18 (range 17–21) and 52 (range 35–86); 30% and 47% positive for CD4 and CCR5, respectively, as determined by flow cytometry), medium (MFI 124 (116–132) and 178 (149–193); 55% and 69%), or high (MFI 773 (689–883) and 516 (459–584); 76% and 78%) CD4 and CCR5. Envs from brain and lymphoid tissue from the same patient were assayed simultaneously on the same populations of transfected Cf2 cells. The well characterized ADA and YU2 Envs cloned from macrophage-tropic blood viral isolates were used as controls in parallel experiments. For most Envs, fusion increased as CD4 and CCR5 levels increased, with greater levels of overall fusion for brain compared with lymphoid Envs at all levels of CD4 and CCR5 (Fig 1A). To determine whether there are differences between brain and lymphoid Envs in their relative use of low CD4 levels, we calculated the ratio for Env-mediated fusion with cells expressing low versus high CD4 (Fig. 1B). MACS2 brain Envs (n=8) fused more efficiently with cells expressing low or medium CD4 levels than did MACS2 lymphoid Envs (n=6) (Fig. 1A). Seven of 8 MACS3 brain Envs and 7 of 8 UK7 brain Envs mediated fusion at low CD4 levels compared with only 1 of 6 MACS3 lymphoid Envs (LN2) and none of the UK7 lymphoid Envs (Fig. 1). In contrast, the ability of UK1 brain versus lymphoid Envs to use low CD4 levels did not segregate according to tissue of origin. Both brain (8/11) and lymphoid (3/6) (SP2, SP6-20, SP6-22) UK1 Envs mediated fusion at low CD4. In total, 30 of 35 brain Envs but only 4 of 20 lymphoid Envs (p<0.01, Fisher’s Exact test) mediated fusion with cells expressing low CD4 levels.
Fig. 1. Cell-cell fusion mediated by brain and lymphoid Envs.
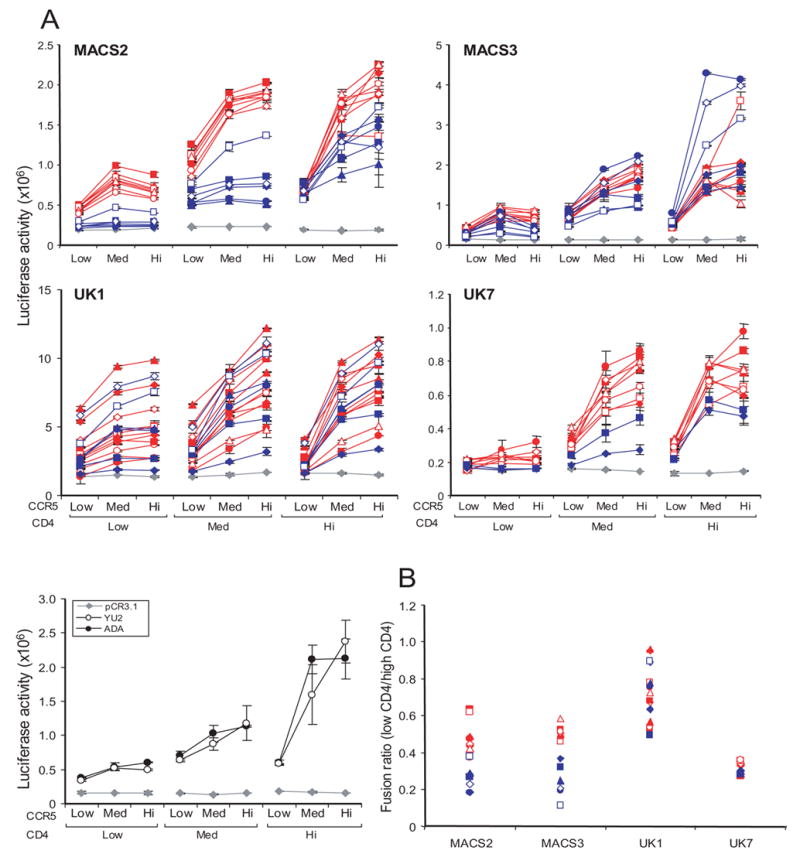
(A) 293T cells expressing brain (red) or lymphoid (blue) Envs from MACS2, MACS3, UK1, and UK7 or the ADA or YU2 Envs were mixed with Cf2luc cells expressing low, medium (Med), or high (Hi) levels of CD4 and CCR5 as indicated. Fusion was measured as luciferase counts in lysed cells following 12 h incubation. Control fusion of 293T cells with pCR3.1 in place of Env, indicating background levels, is represented by grey diamonds. Luciferase activity less than 1.5-fold above the background level obtained with the no Env negative control (pCR3.1) was considered negative. Data are expressed as means of duplicate wells from a single experiment and are representative of results from 2 to 3 independent experiments. Error bars represent standard deviations. (B) Fusion activity of brain (open symbols) and lymphoid (closed symbols) Envs from MACS2, MACS3, UK1 and UK7 expressed as a ratio of Env-mediated fusion with cells expressing low compared to high levels of CD4 and medium levels of CCR5.
In addition to mediating fusion at low CD4 levels, 11 UK1 Envs had an enhanced capacity to induce fusion with cells expressing low CCR5 levels compared to MACS2, MACS3, and UK7 Envs (Fig. 1A). Furthermore, MACS2 and UK7 brain Envs were more efficient than lymphoid Envs at mediating fusion at low CCR5 levels when CD4 was at low or medium levels. In contrast, MACS3 brain Envs were only marginally more efficient than lymphoid Envs at mediating fusion at low CCR5 levels, and only when CD4 levels were also low (Fig. 1A). Therefore, 3 patterns emerged from analyses of Envs from these patients: 1) In MACS2 and UK7, brain Envs were more efficient than lymphoid Envs at mediating fusion at low CD4 and CCR5 levels; 2) In MACS3, brain Envs were more efficient than lymphoid Envs at mediating fusion at low CD4 but not CCR5 levels; and 3) In UK1, Env clones from both brain and lymphoid tissue had a markedly enhanced capacity to mediate fusion at low levels of CD4 and CCR5.
To test the dependence of brain and lymphoid Envs on CD4 and CCR5 levels when mediating virus infection, we performed single round virus entry assays with Cf2 cells expressing low, medium, or high CD4 and CCR5. For these assays, we tested 33 genetically distinct Envs exhibiting a range of abilities to use different CD4 levels similar to that of all Envs cloned from the same tissue sample. Envs that used low CD4 levels in cell-to-cell fusion assays (Fig. 1) also mediated detectable infection of cells expressing low CD4 levels in infection assays (Fig. 2). MACS2, MACS3, and UK7 brain Envs had a greater capacity to infect cells expressing low CD4 than did lymphoid Envs. In contrast, infection mediated by UK1 Envs showed no clear distinction between brain versus lymphoid Envs in the ability to use low CD4, similar to results observed in fusion assays. All UK1 brain and lymphoid Envs, except spleen clone 20, infected cells expressing low CD4 more efficiently than the majority of MACS3, UK1, and UK7 brain or lymphoid Envs (Fig. 2). Brain Envs had higher infectivity ratios for cells expressing low versus high CD4 compared to lymphoid Envs (2p<0.001, Wilcoxon Rank-Sum Test, with combined data stratified by patient). When data from individual patients were analyzed, only MACS3 showed a statistically significant difference between brain and lymphoid Envs (2p=0.016). In contrast, no segregation between brain and lymphoid Envs was observed for infection of cells expressing low versus high levels of CCR5 (data not shown). However, similar to data obtained in cell-cell fusion assays, UK1 Envs from both brain and lymphoid tissue efficiently used low levels of CCR5 to mediate infection, and brain Envs from MACS2 and UK7 used low CCR5 more efficiently than lymphoid Envs (data not shown). Western blot analysis of Env expression demonstrated some variability in levels of gp160 expression and processing (Fig. 3 and data not shown). However, these differences did not correlate with differences in the ability to mediate fusion at low CD4 or CCR5 levels. These results demonstrate that Envs that can use low levels of CD4 and CCR5 for fusion and infection are more frequent in brain compared to lymphoid tissue from the same patient.
Fig. 2. Virus entry mediated by brain and lymphoid Envs.
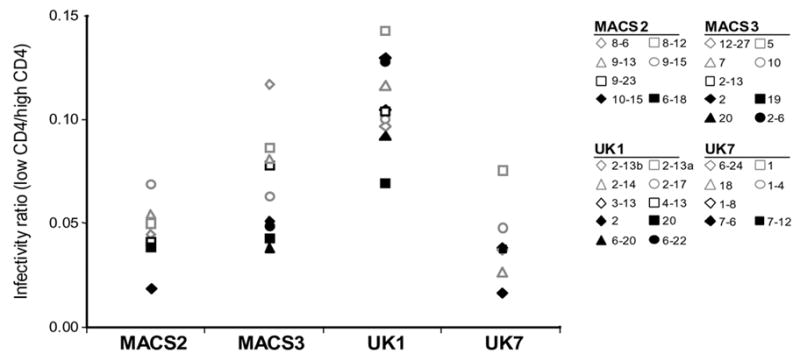
Luciferase reporter viruses were pseudotyped with brain (open symbols) or lymphoid (closed symbols) Envs from MACS2, MACS3, UK1 and UK7. Entry into Cf2 cells is expressed as a ratio of virus entry on cells expressing low compared to high levels of CD4 and high levels of CCR5. Actual values for luciferase counts in cells expressing low or high CD4 ranged from 823-101393, 788-34703, 1053-1072087, and 315-58126 for Envs from MACS2, MACS3, UK1 and UK7, respectively. Background levels were determined as in Fig. 1.
Fig. 3. gp160 and gp120 expression by brain and lymphoid Env clones.
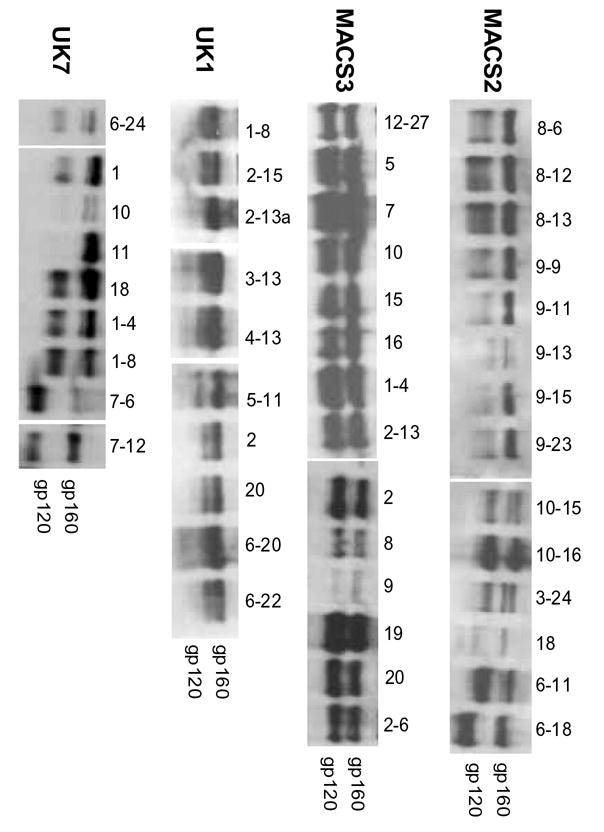
Env expression and processing of gp160 to gp120 in 293T cells detected by Western blot with anti-gp120 rabbit sera. Positions of gp160 and gp120 bands are indicated on the right.
To assess the ability of brain and lymphoid Envs to mediate entry into macrophages, we performed single round entry assays. Equivalent amounts of luciferase reporter viruses pseudotyped with brain or lymphoid Envs were used to infect primary MDM. Both brain and lymphoid Envs mediated a broad range of entry levels into MDM. Levels of Env-mediated entry into MDM did not clearly segregate according to tissue of origin or reduced CD4 dependence. (Fig. 4A). UK1 Envs with a greatly enhanced capacity to use low CD4 and CCR5 levels, SP6-20 and BG3-13, also mediated high levels of entry into MDM, suggesting that their reduced dependence on CD4 and CCR5 levels contributed to their increased M-tropism. Levels of entry into MDM mediated by individual Env clones across the entire data set correlated with their levels of overall fusion activity at low, medium, and high levels of CD4 (Fig. 4B). Together, these results suggest that efficient entry into MDM is dependent on several factors that include the ability of Envs to use low receptor levels and additional factors that influence the overall efficiency and kinetics of fusion.
Fig. 4. Virus entry into macrophages mediated by brain and lymphoid Envs.
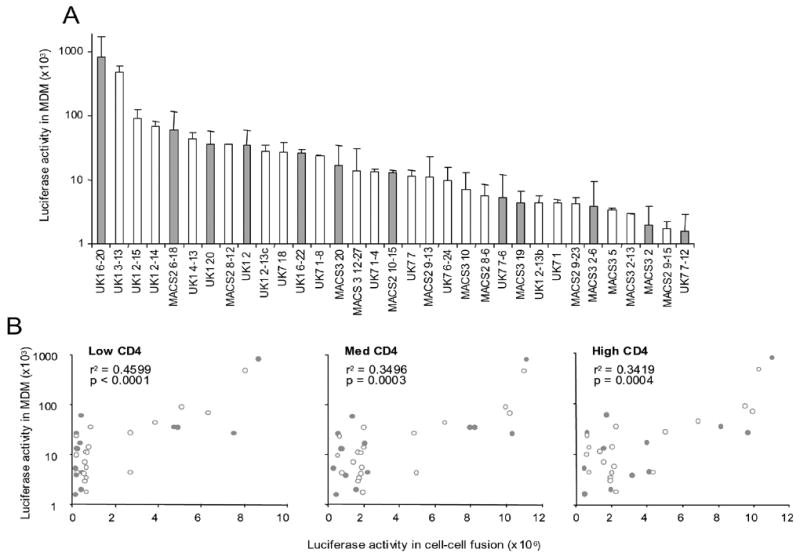
(A) Luciferase reporter viruses were pseudotyped with brain (open bars) or lymphoid (grey bars) Envs from MACS2, MACS3, UK1 and UK7. Entry into macrophages was measured as luciferase counts in lysed cells following incubation for 6 days. Data are expressed as means from duplicate wells and are representative of results in 2 different donors. Error bars represent standard deviations. (B) Brain (open symbols) and lymphoid (grey symbols) entry into MDM (y axis) correlates with levels of fusion between Env-expressing 293T cells and Cf2luc cells expressing low, medium, or high levels of CD4 and high CCR5 (x axis). Pearson's correlation coefficient (r2) and p values are shown.
Analysis of HIV Env sequences
To investigate genetic relationships between env genes in brain and lymphoid tissue, we performed phylogenetic analysis of full-length gp160 nucleotide sequences. No stop codons or frame shifts were observed in the env open reading frames. Env sequences from the MACS2, UK1, and UK7 brain isolates previously obtained by PBMC coculture (Dunfee et al., 2006b; Gorry et al., 2002) were included for comparison. gp160 sequences grouped by patient were most closely related to other sequences from the same patient, with no evidence for possible contaminants (Fig. 5). Distinct env populations according to tissue of origin confirmed genetic partitioning of Env sequences between brain and lymphoid tissue (Fig. 5). Unexpectedly, Env clones from the MACS2 primary brain isolate, which do not have reduced CD4 dependence, clustered with lymphoid rather than brain Envs. In contrast, Env clones from the UK1 and UK7 primary brain isolates, which have reduced CD4 dependence, clustered with Env clones amplified directly from brain tissue. Deletions and insertions in gp41 from the UK1 and MACS2 brain isolate-derived Envs (Gorry et al., 2002) were rare in functional envs amplified directly from tissues, with a truncation near the gp41 C-terminus observed only in UK7 lymphoid clone 7–6. The number of predicted N-linked glycosylation sites in brain and lymphoid Envs ranged from 23 to 28 in gp120, and 4 to 5 in gp41 (Table 2). There was no consistent difference in the number or distribution of predicted N-linked glycans between brain and lymphoid Envs. Screening for hypermutation in brain and lymphoid env genes was performed using HYPERMUT to compare the rate of G to A transitions in each env sequence compared to the tissue consensus (Rose and Korber, 2000). Overall, the hypermutation rate was low (<5%) with no significant difference between brain and lymphoid env genes. However, we excluded nonfunctional Env clones from the data set, which would likely exclude highly mutated Env sequences. The highest frequency of G to A transitions compared to the tissue consensus was seen in 3 UK1 envs, BG3-10, SP5-5 and SP5-11. Overall, these envs were the most genetically distinct compared to other Envs from this patient (Fig. 5). These analyses confirm genetic compartmentalization of full-length, functional Env sequences and underscore the importance of viral genotype and phenotype analysis without prior in vitro amplification.
Fig. 5. Phylogenetic analysis of full-length HIV env nucleotide sequences from autopsy tissue samples and viral isolates.
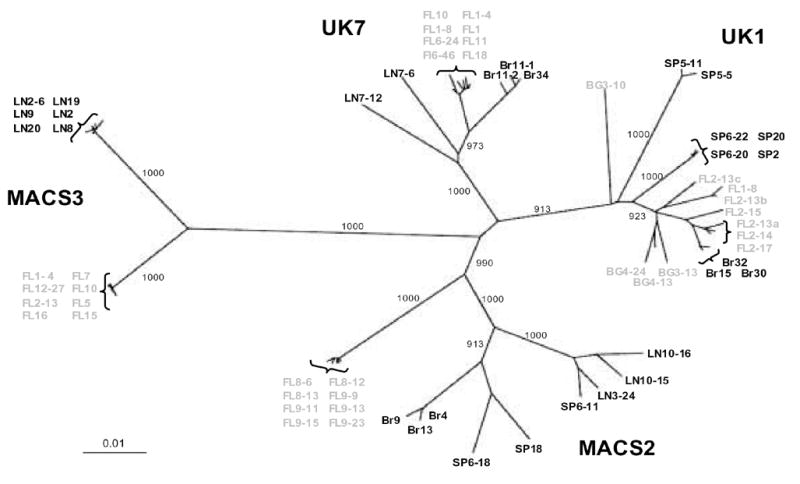
Env sequences amplified directly from brain and lymphoid tissues are color-coded grey and black, respectively. Env sequences for MACS2, UK1 and UK7 brain viral isolates obtained by PBMC co-culture are shown in black with the prefix Br. Numbers associated with each branch are bootstrap values, which represent the number of trees, out of 1000 replicates performed, in which the same branching order was found. Only values above 900 for the major branches are shown. Branch lengths are proportional to the amount of sequence divergence. Scale bars indicate 1% sequence divergence. FL, frontal lobe; BG, basal anglia; LN, lymph node; SP, spleen.
TABLE 2.
Predicted N-linked glycosylation sites in Env clones from brain and lymphoid tissue
| V1 | V2 | V3 | V4 | gp120 | gp41 | ||
|---|---|---|---|---|---|---|---|
| MACS2 | Brain | 1 | 1 | 1 | 5 | 23 | 5 |
| Lymphoid | 1 | 2 | 1 | 5 | 25 | 5 | |
| MACS3 | Brain | 2 | 2 | 1 | 5 | 26 | 5 |
| Lymphoid | 2 | 2 | 1 | 4 | 25 | 4 | |
| UK1 | Brain | 4 | 2 | 1 | 6 | 27 | 5 |
| Lymphoid | 4 | 2 | 1 | 6 | 27 | 5 | |
| UK7 | Brain | 2 | 2 | 1 | 4 | 25 | 4 |
| Lymphoid | 1 | 2 | 1 | 4 | 23 | 4 |
Shown is the median number of predicted N-linked glycosylation sites in all Envs from the indicated tissue.
Proline at position 308 in the V3 loop of HIV gp120 is associated with brain Env
To identify specific amino acid residues that are more frequent in brain compared to lymphoid Envs, we performed a signature pattern analysis of all gp160 amino acid sequences in the data set using VESPA (Korber and Myers, 1992). Proline at position 308, immediately N-terminal of the GPGR sequence in the crown of the V3 loop, was associated with compartmentalization in brain across the entire data set (p<0.01, Fisher’s Exact test). Proline at position 308 was present in 100% (27/27) of brain Envs, but only 28% (4/14) of lymphoid Envs in 3 patients (MACS2, UK1, and UK7) (Fig. 6). All lymphoid Envs with proline at position 308 were from UK1 (SP2, 20, 6–20, and 6–22). All MACS3 Env clones contained histidine at position 308. The clade B consensus at position 308 is histidine, which is found in 53% of clade B V3 sequences, while proline is found at position 308 in only 14% of clade B V3 sequences (n=23,516 (http://hiv.lanl.gov)). In 8 published studies of matched brain and lymphoid or blood samples, proline at position 308 is present in a higher proportion of brain (69/135, 51.1%) compared with lymphoid Envs (45/125, 36.8%) (n=13 subjects that had P308 in any Env, p=0.01, Fisher’s Exact test) (Donaldson et al., 1994; Gartner et al., 1997; Korber et al., 1994; Martin-Garcia et al., 2006; Morris et al., 1999; Peters et al., 2004; van't Wout et al., 1998; Wang et al., 2001). Therefore, proline at position 308 in the V3 region of gp120 is more frequent in brain than in lymphoid tissue.
Fig. 6. Amino acid alignment of HIV Env V3 loop regions of brain and lymphoid Env clones with the clade B consensus.
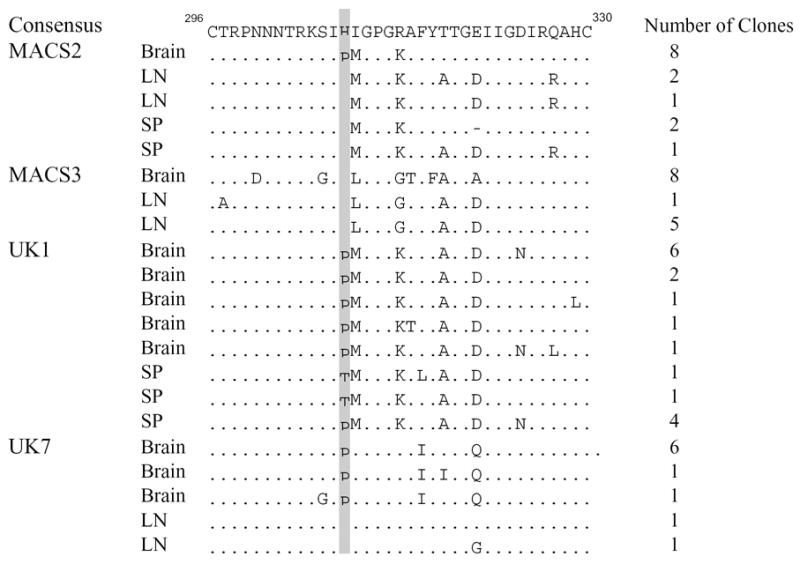
Tissue of origin and number of clones with each sequence are indicated to the left and right of each sequence, respectively. Numbering is relative to the Hxbc2 Env sequence. Dots indicate residues identical to the clade B consensus, and dashes indicate gaps. Proline at position 308 is shaded in grey. LN, lymph node; SP, spleen.
Reduced CD4 and CCR5 dependence of brain Envs is not altered by mutation of P308
To assess the influence of P308 on functional characteristics of brain-derived Envs, mutagenesis was used to change the proline to histidine at this position in the MACS2 (FL8-12) and UK7 (Br34) brain Env clones. We investigated whether introducing this mutation altered the capacity of these Envs to mediate fusion or infection with cells expressing low CD4 or CCR5 levels. The mutant Envs containing H308 were expressed and processed at levels similar to those of the parental Envs (Fig. 7B). There was no significant difference in the efficiency of fusion or infection of Cf2 cells expressing low, medium, or high CD4 or CCR5 when P308 was changed to histidine (Fig. 7A). Additionally, we found no consistent enhancement or reduction of entry into MDM during single round infection assays using MDM from different donors when the H308 mutation was introduced (Fig. 7C). Thus, P308 does not contribute to the reduced CD4 dependence of these 2 brain-derived Envs.
Fig. 7. Fusion and entry of brain Envs with a proline to histidine substitution at position 308.
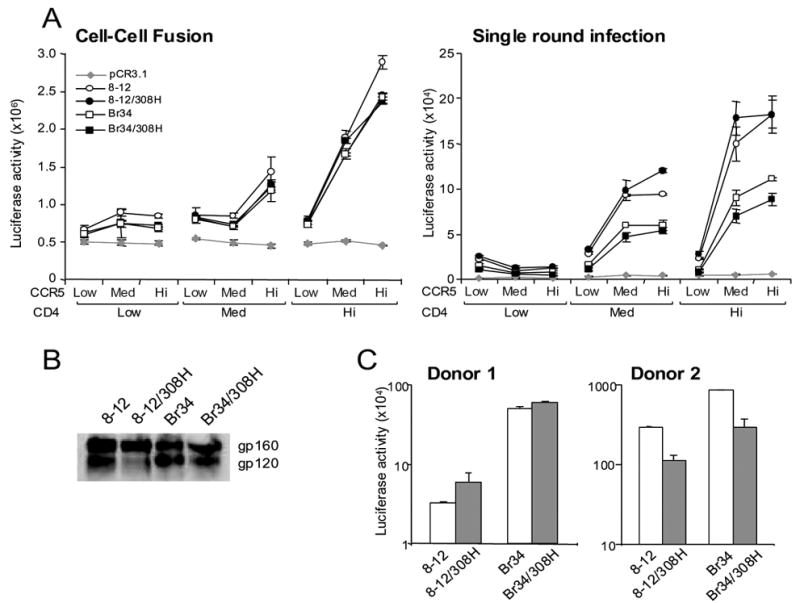
(A) 293T cells or luciferase reporter viruses pseudotyped with wild type and mutant brain Envs from MACS2 (8–12) and UK7 (Br34) were mixed with Cf2luc or Cf2 cells, respectively, expressing low, medium, or high levels of CD4 and CCR5 as indicated. Fusion and infection were measured as luciferase counts in lysed cells following 12 or 72 h incubation, respectively. Background levels were determined as in Fig. 1. (B) Wild type and mutant Env expression and processing of gp160 to gp120 in 293T cells detected by Western blot with anti-gp120 rabbit sera. Positions of gp160 and gp120 bands are indicated on the right. (C) Luciferase reporter viruses pseudotyped with wild type and mutant brain Envs from MACS2 (8–12) and UK7 (Br34) were used to infect MDM from 2 representative donors. Entry was measured as luciferase counts in lysed cells following 6 days incubation. Data are expressed as means from duplicate wells. Error bars represent standard deviations.
Mutation of P308 in brain Envs does not alter sensitivity of brain Envs to entry inhibitors
Mutation of histidine to proline at position 308 in gp120 of a primary HIV isolate was reported in a viral escape mutant that developed resistance to the small molecule CCR5 inhibitor AD101 (Kuhmann et al., 2004; Trkola et al., 2002). The resultant virus was still dependent on CCR5, but contained amino acid substitutions that altered the manner in which Env interacts with CCR5 (Marozsan et al., 2005; Trkola et al., 2002). A change from histidine to proline at position 308 conferred an ~500 fold increase in drug resistance in an Env from one subject (Kuhmann et al., 2004). Therefore, we tested the sensitivity of viruses pseudotyped with Envs containing P308 versus H308 to the CCR5 inhibitors AD101, TAK-779 and the anti-CCR5 MAb 2D7, and the HIV fusion inhibitor T20. MACS2 and UK7 brain Envs showed no difference in sensitivity to these inhibitors when P308 was changed to histidine (Table 3). Therefore, in the context of 2 brain-derived Envs, P308 did not affect sensitivity to CCR5 inhibitors. Thus, the contribution of proline at position 308 to development of resistance to coreceptor inhibitors is context-dependent, and additional determinants may be necessary to generate resistance in genetically different Envs from patients.
TABLE 3.
Neutralization of pseudotyped luciferase viruses by entry inhbitors, MAbs, and HIV-infected patient sera
| AD101a | TAK779a | 2D7b | T20b | b12b | 2F5b | PS1c | PS2c | PS3c | PS4c | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MACS2 | 8-12 | 24.7 | 354 | 0.56 | 0.32 | 5.22 | 12.2 | >50 | 393 | 128 | 131 |
| 8-12/308H | 29.8 | 380 | 0.53 | 0.35 | 4.53 | 10.3 | >50 | 219 | 147 | 200 | |
| UK7 | Br34 | 110.3 | 357 | 3.44 | 0.43 | 0.14 | 10.3 | >50 | 101 | >50 | >50 |
| Br34/308H | 179.7 | 324 | 4.20 | 0.61 | 0.14 | 10.8 | >50 | 143 | >50 | >50 | |
IC50 titers at which luciferase production was reduced by 50%.
nM,
μg/ml,
reciprocal serum dilution
The brain is an immunologically privileged site with reduced selection pressure from cellular and humoral immunity to constrain viral evolution. Viral adaptation to the brain microenvironment can result in enhanced sensitivity to neutralizing antibodies (Martin et al., 2001; Song et al., 2004). Therefore, we tested whether introducing the H308 mutation into the UK7 and MACS2 brain Envs alters sensitivity to neutralization by monoclonal antibodies directed against the CD4 binding site (b12), coreceptor binding site (17b and 19e), or gp41 (2F5), and by sera from 4 HIV-infected individuals. The UK7 brain Env was more sensitive to b12 neutralization, but more resistant to neutralization by patient sera, compared with the MACS2 Env (Table 3). The 17b and 19e MAbs targeting the coreceptor binding site did not reach the IC50 for these Envs at the highest concentration tested (10 μg/ml) in the presence or absence of sCD4 (data not shown). We found no difference in the sensitivity to b12, 17b, 19e, or 2F5 neutralization in these Envs when P308 was changed to histidine. Additionally there was no difference in neutralization sensitivity to sera from 3 HIV positive patients (Table 3). However, MACS2 brain Env containing H308 was more resistant to neutralization by sera from an HIV-infected patient (PS2) compared to the parental Env containing P308, with a 2-fold increase in the IC50 (Table 3). Previous studies have demonstrated a proline substitution at position 308 associated with enhanced neutralization sensitivity (Quinnan et al., 1999; Young et al., 2004; Zhang et al., 2002) suggesting that alteration of this residue in some Envs may increase exposure of neutralization sensitive epitopes. Thus, persistence and compartmentalization of this mutation in the brain may be a consequence of lower neutralizing antibody levels in the CNS compared to the periphery.
Discussion
In this study, we characterized HIV Env clones from brain and lymphoid tissue of 4 AIDS patients with HAD and demonstrated that Envs with reduced dependence on CD4 levels are more frequent in brain compared with lymphoid tissue from the same patient. Thirty of 35 brain Env clones had reduced dependence on CD4 in cell-cell fusion and single round infection assays, while only 4 of 20 lymphoid Envs had reduced CD4 dependence. Previous studies identified HIV Envs with reduced dependence on CD4 levels only in brain (Gorry et al., 2001; Gorry et al., 2002; Martin-Garcia et al., 2006; Peters et al., 2004). Here we report that 4 lymphoid Envs from 2 HAD patients (3 UK1 spleen Envs and 1 MACS3 lymph node Env) have a similar phenotype. Furthermore, 5 brain Envs from UK1, UK7, and MACS3 do not have reduced CD4 dependence. Thus, Envs with reduced CD4 dependence are more frequent in brain, but are also present in lymphoid tissues of some AIDS patients with HAD. These results contrast with previous reports that identified Envs with reduced CD4 dependence only in brain (Martin-Garcia et al., 2006; Peters et al., 2004; Peters et al., 2006), a discrepancy that may be explained in part by the larger number of functional Env clones analyzed in our study. In addition to having reduced CD4 dependence, MACS2 and UK7 brain Envs were also more efficient at utilizing low CCR5 compared with lymphoid Envs. Moreover, a subset of both brain and lymphoid UK1 Envs had a greatly enhanced capacity to use low CCR5 compared to Envs from the other 3 patients. The finding that Envs with reduced CD4 dependence are more frequent in brain than in lymphoid tissues suggests that viral adaptation to the CNS microenvironment selects for viruses with an enhanced capacity to enter cells expressing low levels of these receptors. However, interpatient variability in the capacity of Envs with reduced CD4 dependence to use low CCR5 suggests that complex interdependent relationships exist between CD4 and CCR5 dependence, as has been described for primary HIV isolates (Kozak et al., 1997; Platt et al., 1998).
Reduced CD4 dependence has been associated with increased macrophage tropism of HIV and SIV (Bannert et al., 2000; Gorry et al., 2002; Peters et al., 2004; Puffer et al., 2002). Furthermore, brain Envs with reduced CD4 dependence are macrophage-tropic (Gorry et al., 2002; Peters et al., 2004). We found that UK1 Envs mediated higher levels of entry into MDM than Envs from the other 3 patients, suggesting that their greatly enhanced capacity to use low CD4 and CCR5 levels contributed to their increased M-tropism. However, the ability of Envs across the entire data set to mediate entry into MDM did not show a consistent correlation with reduced dependence on CD4 or CCR5. Rather, the capacity of Envs from both brain and lymphoid tissues to mediate entry into macrophages correlated with overall fusion activity at all levels of CD4 and CCR5. The majority of productive entry into MDM results from virion fusion at the plasma membrane. However, virion internalization in these cells also occurs via macropinocytosis and vesicular uptake into the endocytic pathway (Marechal et al., 1998; Marechal et al., 2001; Schaeffer et al., 2004). HIV entry via the endocytic pathway usually does not lead to productive infection (Marechal et al., 1998). Thus, virus entry via this alternative pathway is unlikely to account for the lack of consistent correlation between MDM entry and reduced CD4 dependence. Together, these findings suggest that the M-tropism of Env clones from brain and lymphoid tissue is dependent not only on the ability to use low receptor levels, but also on additional factors that influence the overall efficiency of fusion. Such factors may include the ability to utilize different isoforms or modified forms of CD4 and CCR5 expressed on MDM (Bannert et al., 2001; Farzan et al., 1999; Lynch et al., 2006; Mummidi et al., 1998), efficiency of binding to cell surface virion attachment factors such as syndecans and type C lectins (Baribaud et al., 2001; Saphire et al., 2001), and the kinetics of gp120 conformational rearrangements and gp41 six-helix bundle formation (Platt et al., 2005; Reeves et al., 2004).
Functional env genes from brain and lymphoid tissue of 4 patients in our study showed distinct tissue specific compartmentalization. However, envs from viruses isolated from brain of MACS2, UK1, and UK7 by coculture with PBMC were distinct from sequences amplified directly from brain tissue, suggesting that viral selection or adaptation occurred during coculture. This was particularly evident for env genes from the MACS2 brain isolate (Br4, Br9 and Br13), which were more closely related to sequences amplified from MACS2 spleen than from brain sequences. The phenotype of the MACS2 virus isolate Envs was also more similar to that of lymphoid Envs from this patient, as these Envs did not exhibit reduced CD4/CCR5 dependence. Similarly, nef genes amplified from the MACS2 brain virus isolate were unlike nef sequences directly amplified from brain tissue, but were similar to nef sequences from lymphoid tissue (Agopian et al., In press). The isolation of this virus from MACS2 brain tissue could be the result of selection in the PBMC coculture of virus present in contaminating blood vessels that contained blood or lymphoid env sequences. UK1 envs were amplified from both frontal lobe and basal ganglia regions of brain. The env genes from these brain regions were genetically distinct, consistent with previous reports of genetically distinct HIV sequences in different regions of brain (Morris et al., 1999; Salemi et al., 2005; Shapshak et al., 1999). The basal ganglia is the brain region most severely affected by HIV encephalitis (Berger and Arendt, 2000; Berger and Nath, 1997; Brew et al., 1995). One basal ganglia Env, BG3-13, exhibited enhanced entry into MDM, but other Envs cloned from this region did not exhibit enhanced macrophage tropism or increased cytopathicity compared to those from matched frontal lobe. Thus, other mechanisms are likely to explain the preferential susceptibility of this region to HIV-induced neurological injury.
In our data set, proline at position 308 in the V3 region of gp120 was associated with brain compartmentalization in 3 patients. P308 was present in 100% of brain Envs, but only 28% of lymphoid Envs from these patients. We also found a higher frequency of P308 in brain (51%; n=135) compared with matched non-brain (37%; n=125) Env sequences when we analyzed data sets from several previous studies (p=0.01, Fisher’s Exact test) (Donaldson et al., 1994; Gartner et al., 1997; Korber et al., 1994; Martin-Garcia et al., 2006; Morris et al., 1999; Peters et al., 2004; van't Wout et al., 1998; Wang et al., 2001). Power et al reported an association between the amino acid residues at positions 308 and 332 with clinical signs of AIDS dementia (Power et al., 1995), but other studies have questioned the role of a specific amino acid at position 308 in neutrotropism (Di Stefano et al., 1996). However, further evidence implicating amino acid alterations at position 308 in viral adaptation to the CNS was recently described in a comparison of CSF and plasma samples (Pillai et al., 2006; Strain et al., 2005). In this study, proline at position 308 was present in Envs from 4 patients with compartmentalized env sequences and proline was significantly correlated with CSF origin (31/43 in CSF and 2/48 in plasma, p<0.001, Fisher’s Exact test) (Pillai et al., 2006). A proline at position 308 was reported in V3 loop sequences from astrocytes but not macrophages/microglia microdissected from brain of one HAD patient (Thompson et al., 2004), raising the question of whether latent HIV within astrocytes may be the source of some sequences amplified in our study. Our data combined with studies of matched CSF and plasma samples imply that proline at position 308 in the V3 loop is a shared signature sequence resulting from adaptation to the brain microenvironment in some HIV-infected individuals.
Position 308 lies at the tip of the V3 loop and thus may be involved in interaction with the chemokine coreceptor. However, it is not a position used for genotyping Envs for CCR5 or CXCR4 usage (Jensen et al., 2003; Resch et al., 2001). The importance of this residue in modulating Env interactions with CCR5 was highlighted by an in vitro generated escape mutant from the CCR5 small molecule inhibitor AD101, with a mutation of histidine to proline at position 308 conferring resistance (Kuhmann et al., 2004; Marozsan et al., 2005; Trkola et al., 2002). However, mutation of P308 to histidine in brain Envs from MACS2 and UK7 did not alter sensitivity to CCR5 inhibitors. This result suggests that escape from coreceptor inhibitors resulting from alterations in single residues in HIV Env is context-dependent.
Molecular modeling of the JRFL gp120-CD4-CD4i Ab X5 complex crystal structure (2B4C) with Swiss PDB Viewer (http://ca.expasy.org/spdbv) suggests that introduction of a proline at position 308 induces minor conformational changes in V3 that result in a slightly more open conformation of the V3 tip region (R. Dunfee and D. Gabuzda, unpublished data). This conformation may allow for increased exposure of the coreceptor binding site or increased exposure of neutralizing epitopes in the N-terminus of V3. Consistent with this model, P308 in conjunction with a methionine at position 309 has been implicated in inducing a more open conformation of HIV Env, resulting in CD4 independence and increased elicitation of broadly neutralizing Abs by a primary HIV Env (Quinnan et al., 1999; Young et al., 2004; Zhang et al., 2002). Envs from MACS2 brain and UK1 brain and spleen contain the P308/M309 motif, but did not mediate CD4-independent infection. However, mutagenesis of P308 to histidine in a MACS2 brain Env decreased sensitivity to neutralizing sera. A microglia-adapted virus, Bori-15, is also more sensitive to serum neutralization (Martin et al., 2001), suggesting that adaptation of brain Envs to the reduced receptor levels on target cells in the brain may result in enhanced exposure of neutralizing epitopes. In the periphery, sensitivity to neutralization by serum antibodies would likely be lethal to an evolving virus. However, the CNS is an immunologically privileged site, with lower neutralizing activity than plasma (Goudsmit et al., 1987; Pillai et al., 2006). Therefore mutations such as P308 may persist in the CNS even if they expose neutralization-sensitive epitopes. Understanding the selective pressures on evolution of the HIV envelope glycoproteins within the brain microenvironment may facilitate not only a better understanding of mechanisms that determine viral tropism for specific target cells and neutralizing epitopes, but also the development of therapeutics to target neurological injury in AIDS patients.
Materials and Methods
Tissue samples
MACS2 and MACS3 were participants in the Chicago component of the Multicenter AIDS Cohort study (MACS) (Gorry et al., 2001; Gorry et al., 2002). Tissue samples from patients UK1 and UK7 were obtained from the Edinburgh MRC HIV Brain and Tissue Bank (Western General Hospital, Edinburgh, UK). Autopsy brain and lymphoid tissue samples were stored at −80°C. Clinical characteristics of patients and tissue samples are shown in Table 1.
Cells
293T, Cf2th (a canine thymocyte cell line) (Choe et al., 1996), and HeLa clone JC53 expressing high levels of CD4 and CCR5 (Platt et al., 1998), provided by D. Kabat), were cultured in Dulbecco's modified Eagle medium supplemented with 10% (vol/vol) fetal bovine serum and 100 μg of penicillin and streptomycin per ml. Cf2-Luc cells (Etemad-Moghadam et al., 2000), derived from Cf2th cells, stably express the firefly luciferase gene under the control of the HIV long terminal repeat and were cultured in medium containing 0.7 mg/ml of G418 (Mediatech, Herndon, VA). Cf2/CD4/R5 cells stably expressing CD4 and CCR5 were cultured in medium containing 0.4 mg/ml of G418, and 0.15 mg/ml hygromycin (Roche, Indianapolis, IN). Monocyte derived macrophages (MDM) were purified from PBMC by plastic adherence and cultured for 5 days in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 μg of penicillin and streptomycin per ml and 10 ng/ml macrophage colony stimulating factor (R&D Systems, Minneapolis, MN.).
PCR amplification, HIV Env cloning, and sequence analysis
Genomic DNA was extracted from tissue samples using Puregene DNA Purification kit (Gentra Systems, Inc., Minneapolis, MN). HIV Gag DNA copy number per 106 cells was determined with TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) using the following primers and probe: 793-gag-F5’GGTGCGAGAGCGTCAGTATTAAG3’, 911-gag-R5’AGCTCCCTGCTTGCCCATA3’ and 835-gag-probe5’TGGGAAAAAATTCGGTTAAGGCCAGGG. Full-length Env genes were amplified from genomic DNA and cloned into pCR3.1 as described (Gorry et al., 2002). Env clones were sequenced using DYEnamic ET Terminator Cycle Sequencing kit (GE Healthcare, Piscataway, NJ) on an Applied Biosystems 3100 Genetic Analyzer (Foster City, CA).
Western blot
Analysis of Env expression was performed as described (Gorry et al., 2002). Briefly, 293T cells were transfected with pCR3.1Env clones and 72 h after transfection were rinsed twice in PBS and resuspended in lysis buffer (Gorry et al., 2002). Equal amounts of cell lysates were run on SDS-PAGE gels and analyzed by western blotting with rabbit anti-gp120 polyclonal sera (provided by J. Sodroski).
Cell-cell fusion assay
Env mediated fusion was quantitated as described previously (Gorry et al., 2002). Briefly, 293T cell cotransfected with pCR3.1Env and pLTR-Tat were mixed with Cf2-Luc cells cotransfected with 0.05 (low), 0.5 (med), or 5 μg (hi) amounts of pcDNA3-CD4 and pcDNA3-CCR5 resulting in low, medium, or high cell surface expression of CD4 and CCR5 as determined by flow cytometry. Fourteen to 16 h later, cells were harvested and assayed for luciferase activity (Promega, Madison, WI).
Single round infection with pseudotyped viruses
To screen for functional Env clones, 293T cells were cotransfected with pCR3.1Env and pNL4.3 envGFP. GFP pseudoviruses were harvested 72 h after transfection and used to infect JC53 cells. GFP positive cells were detected by fluorescence microscopy 48-72 h after infection. Luciferase pseudotyped viruses were produced by cotransfection of 293T cells with pCR3.1Env clones and pNL4.3 envLuc. Luciferase pseudoviruses were harvested 72 h after transfection and used to infect Cf2 cells transfected with CD4 and either CCR5, CXCR4, or CCR3. 72 h later, cells were harvested and assayed for luciferase activity. MDM were harvested and assayed for luciferase activity 6 days after infection with luciferase pseudoviruses.
Virus inhibition and neutralization assays
CCR5 inhibitors AD101 (SCH 350581, provided by Schering-Plough Research Institute, Kenilworth, NJ.), TAK-779, and 2D7 (both obtained through the AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, NIH, Bethesda, MD.) were preincubated with Cf2/CD4/R5 target cells for 1 h prior to infection with luciferase pseudoviruses. For T20 (from Roche, obtained through the AIDS Research and Reference Reagents Program) and human monoclonal Abs against HIV Env, virus was incubated with for 1 h with a range of concentrations of T20 or each MAb prior to infection of Cf2/CD4/R5 cells. 72 h later, cells were harvested and assayed for luciferase activity. MAb b12 (Roben et al., 1994) and 2F5 (Buchacher et al., 1994; Purtscher et al., 1994) were obtained though the AIDS Research and Reference Reagents Program (from D. Burton and C. Barbas, and Herman Katinger respectively). MAbs 17b (Thali et al., 1993) and 19e (Decker et al., 2005) were kindly provided by J. Robinson (Tulane University Health Sciences Center, New Orleans, La.). HIV-position patient sera, PS1 and PS2 (Vujcic and Quinnan, 1995), were obtained though the AIDS Research and Reference Reagents Program from Dr. Luba Vujcic. PS3 and PS4 were kindly provided by J. Sodroski.
Phylogenetic analysis
Sequences were aligned using VectorNTI (Invitrogen, Carlsbad, CA) and bootstrapped phylogenetic trees were created by the neighbor joining method using ClustalX (Thompson et al., 1997). Trees were visualized using Treeview (Page, 1996). Potential N-linked glycosylation sites were predicted using N-Glycosite (http://hiv-web.lanl.gov). Sequence signatures in brain and lymphoid sequence sets were identified using VESPA (http://hiv-web.lanl.gov) (Korber and Myers, 1992).
Nucleotide sequence accession numbers
Genbank accession numbers for gp160 nucleotide and amino acid sequences are DQ976380-DQ976434. gp160 sequences from the UK1, UK7 and MACS2 brain viral isolates have been reported previously (Dunfee et al., 2006b; Gorry et al., 2002). (UK1Br15: AF491740; UK1Br30: AF491741; UK1Br32: AF491742; MACS2Br4: AF491738; MACS2Br9: AF491739; MACS2Br13: AF491737).
Acknowledgments
We thank J. Sodroski, B. Korber, P. Gorry, J. Moore, P. Ancuta, N. Madani, K. Agopian, and J. Wang for helpful discussions and advice, and S. Lee for statistical analysis. We also thank D. Kabat for providing JC53 cells, J. Sodroski for rabbit anti-gp120 sera, PS3, and PS4, J. Robinson for MAbs 17b and 19e, the NIH AIDS Reagent Program for TAK-779, 2D7, b12, 2F5, PS1 and PS2, and J Strizki of Schering-Plough Research Institute for AD101. This work was supported by NIH NS37277. Core facilities were supported by Harvard Medical School Center for AIDS Research (CFAR) and DFCI/Harvard Cancer Center grants. The Edinburgh HIV Brain and Tissue Bank is supported by a grant from MRC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agopian K, Wei BL, Garcia JV, Gabuzda D. CD4 and MHC-I downregulation are conserved in primary HIV-1 Nef alleles from brain and lymphoid tissues, but Pak2 activation is highly variable. Virology. doi: 10.1016/j.virol.2006.07.053. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SF, Groves M, Gray F, Scaravilli F. Early entry and widespread cellular involvement of HIV-1 DNA in brains of HIV-1 positive asymptomatic individuals. J Neuropathol Exp Neurol. 1999;58(11):1156–62. doi: 10.1097/00005072-199911000-00005. [DOI] [PubMed] [Google Scholar]
- Ancuta P, Bakri Y, Chomont N, Hocini H, Gabuzda D, Haeffner-Cavaillon N. Opposite effects of IL-10 on the ability of dendritic cells and macrophages to replicate primary CXCR4-dependent HIV-1 strains. J Immunol. 2001;166(6):4244–53. doi: 10.4049/jimmunol.166.6.4244. [DOI] [PubMed] [Google Scholar]
- Bannert N, Craig S, Farzan M, Sogah D, Santo NV, Choe H, Sodroski J. Sialylated O-glycans and sulfated tyrosines in the NH2-terminal domain of CC chemokine receptor 5 contribute to high affinity binding of chemokines. J Exp Med. 2001;194(11):1661–73. doi: 10.1084/jem.194.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74(23):10984–93. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baribaud F, Pohlmann S, Doms RW. The role of DC-SIGN and DC-SIGNR in HIV and SIV attachment, infection, and transmission. Virology. 2001;286(1):1–6. doi: 10.1006/viro.2001.0975. [DOI] [PubMed] [Google Scholar]
- Berger JR, Arendt G. HIV dementia: the role of the basal ganglia and dopaminergic systems. J Psychopharmacol. 2000;14(3):214–21. doi: 10.1177/026988110001400304. [DOI] [PubMed] [Google Scholar]
- Berger JR, Nath A. HIV dementia and the basal ganglia. Intervirology. 1997;40(2–3):122–31. doi: 10.1159/000150539. [DOI] [PubMed] [Google Scholar]
- Brew BJ, Rosenblum M, Cronin K, Price RW. AIDS dementia complex and HIV-1 brain infection: clinical-virological correlations. Ann Neurol. 1995;38(4):563–70. doi: 10.1002/ana.410380404. [DOI] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses. 1994;10(4):359–69. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Chang J, Jozwiak R, Wang B, Ng T, Ge YC, Bolton W, Dwyer DE, Randle C, Osborn R, Cunningham AL, Saksena NK. Unique HIV type 1 V3 region sequences derived from six different regions of brain: region-specific evolution within host-determined quasispecies. AIDS Res Hum Retroviruses. 1998;14(1):25–30. doi: 10.1089/aid.1998.14.25. [DOI] [PubMed] [Google Scholar]
- Cheng-Mayer C, Liu R, Landau NR, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71(2):1657–61. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85(7):1135–48. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- Cunningham AL, Li S, Juarez J, Lynch G, Alali M, Naif H. The level of HIV infection of macrophages is determined by interaction of viral and host cell genotypes. J Leukoc Biol. 2000;68(3):311–7. [PubMed] [Google Scholar]
- d'Arminio Monforte A, Cinque P, Mocroft A, Goebel FD, Antunes F, Katlama C, Justesen US, Vella S, Kirk O, Lundgren J. Changing incidence of central nervous system diseases in the EuroSIDA cohort. Ann Neurol. 2004;55(3):320–8. doi: 10.1002/ana.10827. [DOI] [PubMed] [Google Scholar]
- Davis LE, Hjelle BL, Miller VE, Palmer DL, Llewellyn AL, Merlin TL, Young SA, Mills RG, Wachsman W, Wiley CA. Early viral brain invasion in iatrogenic human immunodeficiency virus infection. Neurology. 1992;42(9):1736–9. doi: 10.1212/wnl.42.9.1736. [DOI] [PubMed] [Google Scholar]
- Decker JM, Bibollet-Ruche F, Wei X, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med. 2005;201(9):1407–19. doi: 10.1084/jem.20042510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- Di Marzio P, Tse J, Landau NR. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res Hum Retroviruses. 1998;14(2):129–38. doi: 10.1089/aid.1998.14.129. [DOI] [PubMed] [Google Scholar]
- Di Stefano M, Wilt S, Gray F, Dubois-Dalcq M, Chiodi F. HIV type 1 V3 sequences and the development of dementia during AIDS. AIDS Res Hum Retroviruses. 1996;12(6):471–6. doi: 10.1089/aid.1996.12.471. [DOI] [PubMed] [Google Scholar]
- Dick AD, Pell M, Brew BJ, Foulcher E, Sedgwick JD. Direct ex vivo flow cytometric analysis of human microglial cell CD4 expression: examination of central nervous system biopsy specimens from HIV-seropositive patients and patients with other neurological disease. Aids. 1997;11(14):1699–708. doi: 10.1097/00002030-199714000-00006. [DOI] [PubMed] [Google Scholar]
- Dittmar MT, McKnight A, Simmons G, Clapham PR, Weiss RA, Simmonds P. HIV-1 tropism and co-receptor use. Nature. 1997;385(6616):495–6. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- Donaldson YK, Bell JE, Holmes EC, Hughes ES, Brown HK, Simmonds P. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J Virol. 1994;68(9):5991–6005. doi: 10.1128/jvi.68.9.5991-6005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore GJ, Correll PK, Li Y, Kaldor JM, Cooper DA, Brew BJ. Changes to AIDS dementia complex in the era of highly active antiretroviral therapy. Aids. 1999;13(10):1249–53. doi: 10.1097/00002030-199907090-00015. [DOI] [PubMed] [Google Scholar]
- Dore GJ, McDonald A, Li Y, Kaldor JM, Brew BJ. Marked improvement in survival following AIDS dementia complex in the era of highly active antiretroviral therapy. Aids. 2003;17(10):1539–45. doi: 10.1097/00002030-200307040-00015. [DOI] [PubMed] [Google Scholar]
- Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667–73. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- Dunfee RL, Thomas ER, Gorry PR, Wang J, Ancuta P, Gabuzda D. Mechanisms of HIV-1 neurotropism. Curr HIV Res. 2006a;4(3):267–78. doi: 10.2174/157016206777709500. [DOI] [PubMed] [Google Scholar]
- Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A. 2006b doi: 10.1073/pnas.0605513103. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Sun Y, Nicholson EK, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol. 2000;74(9):4433–40. doi: 10.1128/jvi.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–76. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- Fear WR, Kesson AM, Naif H, Lynch GW, Cunningham AL. Differential tropism and chemokine receptor expression of human immunodeficiency virus type 1 in neonatal monocytes, monocyte-derived macrophages, and placental macrophages. J Virol. 1998;72(2):1334–44. doi: 10.1128/jvi.72.2.1334-1344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner S, McDonald RA, Hunter EA, Bouwman F, Liu Y, Popovic M. Gp120 sequence variation in brain and in T-lymphocyte human immunodeficiency virus type 1 primary isolates. J Hum Virol. 1997;1(1):3–18. [PubMed] [Google Scholar]
- Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5(1):69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ, Bell JE, Bannert N, Crawford K, Wang H, Schols D, De Clercq E, Kunstman K, Wolinsky SM, Gabuzda D. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol. 2001;75(21):10073–89. doi: 10.1128/JVI.75.21.10073-10089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76(12):6277–92. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsmit J, Epstein LG, Paul DA, van der Helm HJ, Dawson GJ, Asher DM, Yanagihara R, Wolff AV, Gibbs CJ, Jr, Gajdusek DC. Intra-blood-brain barrier synthesis of human immunodeficiency virus antigen and antibody in humans and chimpanzees. Proc Natl Acad Sci U S A. 1987;84(11):3876–80. doi: 10.1073/pnas.84.11.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani-Bowering GM, Filion LG. Down regulation of CD4 expression following isolation and culture of human monocytes. Clin Diagn Lab Immunol. 2000;7(2):182–91. doi: 10.1128/cdli.7.2.182-191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ES, Bell JE, Simmonds P. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J Virol. 1997;71(2):1272–80. doi: 10.1128/jvi.71.2.1272-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CS, Pontow S, Ratner L. Relationship between productive HIV-1 infection of macrophages and CCR5 utilization. Virology. 1999;264(2):278–88. doi: 10.1006/viro.1999.0013. [DOI] [PubMed] [Google Scholar]
- Jensen MA, Li FS, van 't Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J Virol. 2003;77(24):13376–88. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CA, Watkins BA, Kufta C, Dubois-Dalcq M. Infection of brain microglial cells by human immunodeficiency virus type 1 is CD4 dependent. J Virol. 1991;65(2):736–42. doi: 10.1128/jvi.65.2.736-742.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410(6831):988–94. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005;12(Suppl 1):878–92. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- Kazazi F, Mathijs JM, Foley P, Cunningham AL. Variations in CD4 expression by human monocytes and macrophages and their relationships to infection with the human immunodeficiency virus. J Gen Virol. 1989;70 ( Pt 10):2661–72. doi: 10.1099/0022-1317-70-10-2661. [DOI] [PubMed] [Google Scholar]
- Koning FA, Schols D, Schuitemaker H. No selection for CCR5 coreceptor usage during parenteral transmission of macrophagetropic syncytium-inducing human immunodeficiency virus type 1. J Virol. 2001;75(18):8848–53. doi: 10.1128/JVI.75.18.8848-8853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8(9):1549–60. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, Levy R, Wolinsky SM. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68(11):7467–81. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak SL, Platt EJ, Madani N, Ferro FE, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71(2):873–82. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74(15):7005–15. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhmann SE, Pugach P, Kunstman KJ, Taylor J, Stanfield RL, Snyder A, Strizki JM, Riley J, Baroudy BM, Wilson IA, Korber BT, Wolinsky SM, Moore JP. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J Virol. 2004;78(6):2790–807. doi: 10.1128/JVI.78.6.2790-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin SR, Sonza S, Irving LB, McDonald CF, Mills J, Crowe SM. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res Hum Retroviruses. 1996;12(10):877–83. doi: 10.1089/aid.1996.12.877. [DOI] [PubMed] [Google Scholar]
- Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif HM. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73(12):9741–55. doi: 10.1128/jvi.73.12.9741-9755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch GW, Turville S, Carter B, Sloane AJ, Chan A, Muljadi N, Li S, Low L, Armati P, Raison R, Zoellner H, Williamson P, Cunningham A, Church WB. Marked differences in the structures and protein associations of lymphocyte and monocyte CD4: resolution of a novel CD4 isoform. Immunol Cell Biol. 2006;84(2):154–65. doi: 10.1111/j.1440-1711.2005.01403.x. [DOI] [PubMed] [Google Scholar]
- Marechal V, Clavel F, Heard JM, Schwartz O. Cytosolic Gag p24 as an index of productive entry of human immunodeficiency virus type 1. J Virol. 1998;72(3):2208–12. doi: 10.1128/jvi.72.3.2208-2212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal V, Prevost MC, Petit C, Perret E, Heard JM, Schwartz O. Human immunodeficiency virus type 1 entry into macrophages mediated by macropinocytosis. J Virol. 2001;75(22):11166–77. doi: 10.1128/JVI.75.22.11166-11177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozsan AJ, Kuhmann SE, Morgan T, Herrera C, Rivera-Troche E, Xu S, Baroudy BM, Strizki J, Moore JP. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D) Virology. 2005;338(1):182–99. doi: 10.1016/j.virol.2005.04.035. [DOI] [PubMed] [Google Scholar]
- Martin J, LaBranche CC, Gonzalez-Scarano F. Differential CD4/CCR5 Utilization, gp120 Conformation, and Neutralization Sensitivity between Envelopes from a Microglia-Adapted Human Immunodeficiency Virus Type 1 and Its Parental Isolate. J Virol. 2001;75(8):3568–80. doi: 10.1128/JVI.75.8.3568-3580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology. 2006;346(1):169–79. doi: 10.1016/j.virol.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Martin-Garcia J, Kolson DL, Gonzalez-Scarano F. Chemokine receptors in the brain: their role in HIV infection and pathogenesis. Aids. 2002;16(13):1709–30. doi: 10.1097/00002030-200209060-00003. [DOI] [PubMed] [Google Scholar]
- McArthur JC, Haughey N, Gartner S, Conant K, Pardo C, Nath A, Sacktor N. Human immunodeficiency virus-associated dementia: an evolving disease. J Neurovirol. 2003;9(2):205–21. doi: 10.1080/13550280390194109. [DOI] [PubMed] [Google Scholar]
- Morris A, Marsden M, Halcrow K, Hughes ES, Brettle RP, Bell JE, Simmonds P. Mosaic structure of the human immunodeficiency virus type 1 genome infecting lymphoid cells and the brain: evidence for frequent in vivo recombination events in the evolution of regional populations. J Virol. 1999;73(10):8720–31. doi: 10.1128/jvi.73.10.8720-8731.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummidi S, Ahuja SS, Gonzalez E, Anderson SA, Santiago EN, Stephan KT, Craig FE, O'Connell P, Tryon V, Clark RA, Dolan MJ, Ahuja SK. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4(7):786–93. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- Naif HM, Cunningham AL, Alali M, Li S, Nasr N, Buhler MM, Schols D, de Clercq E, Stewart G. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Delta32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J Virol. 2002;76(7):3114–24. doi: 10.1128/JVI.76.7.3114-3124.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenburg JK, Brodt HR, Herndier BG, Bickel M, Bacchetti P, Price RW, Grant RM, Schlote W. HIV-related neuropathology, 1985 to 1999: rising prevalence of HIV encephalopathy in the era of highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2002;31(2):171–7. doi: 10.1097/00126334-200210010-00007. [DOI] [PubMed] [Google Scholar]
- Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, Schwartz O, Heard JM, Clark-Lewis I, Legler DF, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382(6594):833–5. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B, Taylor J, Levy R, Murphy RL, Wolinsky SM, Gabuzda D. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol. 2003;77(22):12336–45. doi: 10.1128/JVI.77.22.12336-12345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78(13):6915–26. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom c, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, Clapham PR. Non-macrophage-tropic R5 Envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J Virol. 2006 doi: 10.1128/JVI.02328-05. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peudenier S, Hery C, Montagnier L, Tardieu M. Human microglial cells: characterization in cerebral tissue and in primary culture, and study of their susceptibility to HIV-1 infection. Ann Neurol. 1991a;29(2):152–61. doi: 10.1002/ana.410290207. [DOI] [PubMed] [Google Scholar]
- Peudenier S, Hery C, Ng KH, Tardieu M. HIV receptors within the brain: a study of CD4 and MHC-II on human neurons, astrocytes and microglial cells. Res Virol. 1991b;142(2–3):145–9. doi: 10.1016/0923-2516(91)90051-4. [DOI] [PubMed] [Google Scholar]
- Pillai SK, Kosakovsky Pond SL, Liu Y, Good BM, Strain MC, Ellis RJ, Letendre S, Smith DM, Gunthard HF, Grant I, Marcotte TD, Allen McCutchan J, Richman DD, Wong JK. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain. 2006 doi: 10.1093/brain/awl136. in press. [DOI] [PubMed] [Google Scholar]
- Platt EJ, Durnin JP, Kabat D. Kinetic factors control efficiencies of cell entry, efficacies of entry inhibitors, and mechanisms of adaptation of human immunodeficiency virus. J Virol. 2005;79(7):4347–56. doi: 10.1128/JVI.79.7.4347-4356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(4):2855–64. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, McArthur JC, Johnson RT, Griffin DE, Glass JD, Dewey R, Chesebro B. Distinct HIV-1 env sequences are associated with neurotropism and neurovirulence. Curr Top Microbiol Immunol. 1995;202:89–104. doi: 10.1007/978-3-642-79657-9_7. [DOI] [PubMed] [Google Scholar]
- Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol. 2002;76(6):2595–605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, Tauer C, Berger R, Barrett N, Jungbauer A, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1994;10(12):1651–8. doi: 10.1089/aid.1994.10.1651. [DOI] [PubMed] [Google Scholar]
- Quinnan GV, Jr, Zhang PF, Fu DW, Dong M, Alter HJ. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res Hum Retroviruses. 1999;15(6):561–70. doi: 10.1089/088922299311088. [DOI] [PubMed] [Google Scholar]
- Rana S, Besson G, Cook DG, Rucker J, Smyth RJ, Yi Y, Turner JD, Guo HH, Du JG, Peiper SC, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms RW, Parmentier M, Collman RG. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the delta ccr5 mutation. J Virol. 1997;71(4):3219–27. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy RT, Achim CL, Sirko DA, Tehranchi S, Kraus FG, Wong-Staal F, Wiley CA. Sequence analysis of the V3 loop in brain and spleen of patients with HIV encephalitis. AIDS Res Hum Retroviruses. 1996;12(6):477–82. doi: 10.1089/aid.1996.12.477. [DOI] [PubMed] [Google Scholar]
- Reeves JD, Miamidian JL, Biscone MJ, Lee FH, Ahmad N, Pierson TC, Doms RW. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol. 2004;78(10):5476–85. doi: 10.1128/JVI.78.10.5476-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W, Hoffman N, Swanstrom R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology. 2001;288(1):51–62. doi: 10.1006/viro.2001.1087. [DOI] [PubMed] [Google Scholar]
- Reynes J, Portales P, Segondy M, Baillat V, Andre P, Avinens O, Picot MC, Clot J, Eliaou JF, Corbeau P. CD4 T cell surface CCR5 density as a host factor in HIV-1 disease progression. Aids. 2001;15(13):1627–34. doi: 10.1097/00002030-200109070-00004. [DOI] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF, 3rd, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J Virol. 1994;68(8):4821–8. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics. 2000;16(4):400–1. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Sacktor N, Lyles RH, Skolasky R, Kleeberger C, Selnes OA, Miller EN, Becker JT, Cohen B, McArthur JC. HIV-associated neurologic disease incidence changes:: Multicenter AIDS Cohort Study, 1990–1998. Neurology. 2001;56(2):257–60. doi: 10.1212/wnl.56.2.257. [DOI] [PubMed] [Google Scholar]
- Sacktor N, McDermott MP, Marder K, Schifitto G, Selnes OA, McArthur JC, Stern Y, Albert S, Palumbo D, Kieburtz K, De Marcaida JA, Cohen B, Epstein L. HIV-associated cognitive impairment before and after the advent of combination therapy. J Neurovirol. 2002;8(2):136–42. doi: 10.1080/13550280290049615. [DOI] [PubMed] [Google Scholar]
- Salemi M, Lamers SL, Yu S, de Oliveira T, Fitch WM, McGrath MS. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J Virol. 2005;79(17):11343–52. doi: 10.1128/JVI.79.17.11343-11352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire AC, Bobardt MD, Zhang Z, David G, Gallay PA. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J Virol. 2001;75(19):9187–200. doi: 10.1128/JVI.75.19.9187-9200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer E, Soros VB, Greene WC. Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J Virol. 2004;78(3):1375–83. doi: 10.1128/JVI.78.3.1375-1383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapshak P, Segal DM, Crandall KA, Fujimura RK, Zhang BT, Xin KQ, Okuda K, Petito CK, Eisdorfer C, Goodkin K. Independent evolution of HIV type 1 in different brain regions. AIDS Res Hum Retroviruses. 1999;15(9):811–20. doi: 10.1089/088922299310719. [DOI] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, McKnight A, Dejucq N, Hibbitts S, Power CA, Aarons E, Schols D, De Clercq E, Proudfoot AE, Clapham PR. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72(10):8453–7. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Yi Y, Isaacs SN, Kolson DL, Collman RG. Concordant utilization of macrophage entry coreceptors by related variants within an HIV type 1 primary isolate viral swarm. AIDS Res Hum Retroviruses. 2001;17(10):957–63. doi: 10.1089/088922201750290078. [DOI] [PubMed] [Google Scholar]
- Song B, Cayabyab M, Phan N, Wang L, Axthelm MK, Letvin NL, Sodroski JG. Neutralization sensitivity of a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2N) isolated from an infected rhesus macaque with neurological disease. Virology. 2004;322(1):168–81. doi: 10.1016/j.virol.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Strain MC, Letendre S, Pillai SK, Russell T, Ignacio CC, Gunthard HF, Good B, Smith DM, Wolinsky SM, Furtado M, Marquie-Beck J, Durelle J, Grant I, Richman DD, Marcotte T, McCutchan JA, Ellis RJ, Wong JK. Genetic composition of human immunodeficiency virus type 1 in cerebrospinal fluid and blood without treatment and during failing antiretroviral therapy. J Virol. 2005;79(3):1772–88. doi: 10.1128/JVI.79.3.1772-1788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67(7):3978–88. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–82. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KA, Churchill MJ, Gorry PR, Sterjovski J, Oelrichs RB, Wesselingh SL, McLean CA. Astrocyte specific viral strains in HIV dementia. Ann Neurol. 2004;56(6):873–7. doi: 10.1002/ana.20304. [DOI] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384(6605):184–7. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Trkola A, Kuhmann SE, Strizki JM, Maxwell E, Ketas T, Morgan T, Pugach P, Xu S, Wojcik L, Tagat J, Palani A, Shapiro S, Clader JW, McCombie S, Reyes GR, Baroudy BM, Moore JP. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc Natl Acad Sci U S A. 2002;99(1):395–400. doi: 10.1073/pnas.012519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle DL, Harrison JK, Anders C, Sleasman JW, Goodenow MM. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol. 1998;72(6):4962–9. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcour V, Shikuma C, Shiramizu B, Watters M, Poff P, Selnes O, Holck P, Grove J, Sacktor N. Higher frequency of dementia in older HIV-1 individuals: the Hawaii Aging with HIV-1 Cohort. Neurology. 2004;63(5):822–7. doi: 10.1212/01.wnl.0000134665.58343.8d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van't Wout AB, Ran LJ, Kuiken CL, Kootstra NA, Pals ST, Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72(1):488–96. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujcic LK, Quinnan GV., Jr Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res Hum Retroviruses. 1995;11(7):783–7. doi: 10.1089/aid.1995.11.783. [DOI] [PubMed] [Google Scholar]
- Wang J, Crawford K, Yuan M, Wang H, Gorry PR, Gabuzda D. Regulation of CC chemokine receptor 5 and CD4 expression and human immunodeficiency virus type 1 replication in human macrophages and microglia by T helper type 2 cytokines. J Infect Dis. 2002;185(7):885–97. doi: 10.1086/339522. [DOI] [PubMed] [Google Scholar]
- Wang TH, Donaldson YK, Brettle RP, Bell JE, Simmonds P. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J Virol. 2001;75(23):11686–99. doi: 10.1128/JVI.75.23.11686-11699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Bar-Or A, Ulvestad E, Olivier A, Antel JP, Yong VW. Biology of adult human microglia in culture: comparisons with peripheral blood monocytes and astrocytes. J Neuropathol Exp Neurol. 1992;51(5):538–49. doi: 10.1097/00005072-199209000-00009. [DOI] [PubMed] [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71(3):2059–71. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384(6605):179–83. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- Yi Y, Isaacs SN, Williams DA, Frank I, Schols D, De Clercq E, Kolson DL, Collman RG. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J Virol. 1999;73(9):7117–25. doi: 10.1128/jvi.73.9.7117-7125.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Rana S, Turner JD, Gaddis N, Collman RG. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72(1):772–7. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KR, Teal BE, Brooks Y, Green TD, Bower JF, Ross TM. Unique V3 loop sequence derived from the R2 strain of HIV-type 1 elicits broad neutralizing antibodies. AIDS Res Hum Retroviruses. 2004;20(11):1259–68. doi: 10.1089/0889222042545063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV., Jr A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J Virol. 2002;76(2):644–55. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


