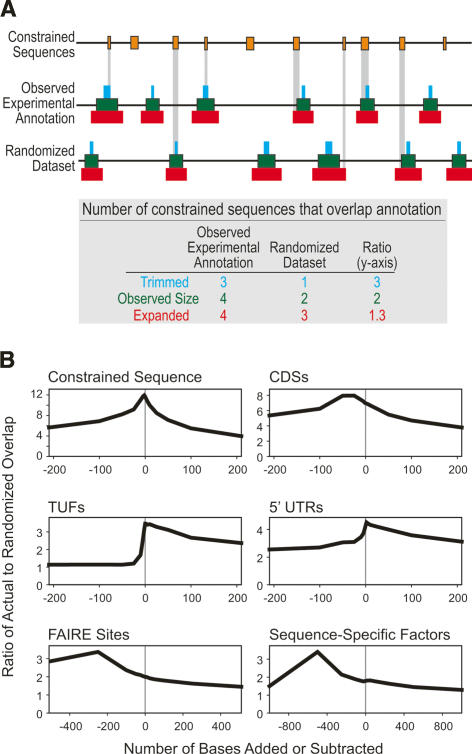
Figure 8.
Significance of constrained sequence overlapping various experimental annotations. We quantified the ratio of “observed” to “randomized” overlaps between constrained sequences and experimental annotations (see Supplemental Box S1), after adding and subtracting a given number of bases to the ends of each experimentally identified annotation. Randomized data sets were generated by randomizing the start positions of features within each ENCODE target, preserving the length distribution of each feature set and any target-specific regional effects. (A) This analysis is illustrated for a hypothetical set of annotations. (Orange bars) The positions of constrained sequences; trimmed (blue bars), observed (green bars), and expanded (red) experimental annotations. (Vertical gray bars) Regions of overlap between constrained sequences and experimental annotations. A table summarizing the overlaps among the different scenarios is provided below the diagram. For this hypothetical example, note how the ratio of overlap between the observed and randomized data sets increases as the experimental annotations are trimmed, indicating an enrichment of constrained sequence in the trimmed annotations. (B) This analysis for several experimentally identified elements is plotted, where the X-axis indicates the amount of trimmed (negative) or expanded (positive) sequence on each element, and the Y-axis indicates the ratio of observed-to-randomized overlap (scale varies between plots). Note that CDSs exhibit a slight enrichment after deletion of a small number of bases at either end, but are very similar to what is expected given the theoretically optimal self–self overlap (“Constrained Sequence”), where we know that trimming should not increase specificity. For many annotations (e.g., “TUFs” and “5′-UTRs”) (see Supplemental Box S1), such enrichment quickly drops off as the annotations are expanded or trimmed. However, some annotations, such as “FAIRE Sites” and “Sequence-Specific Factors,” exhibit a clear improvement in overlap after trimming substantial amounts of sequence from either end (250 and 500 bases for “FAIRE Sites” and “Sequence-Specific Factors,” respectively). Similar plots for all experimental annotations are available as Supplemental Figure S4.