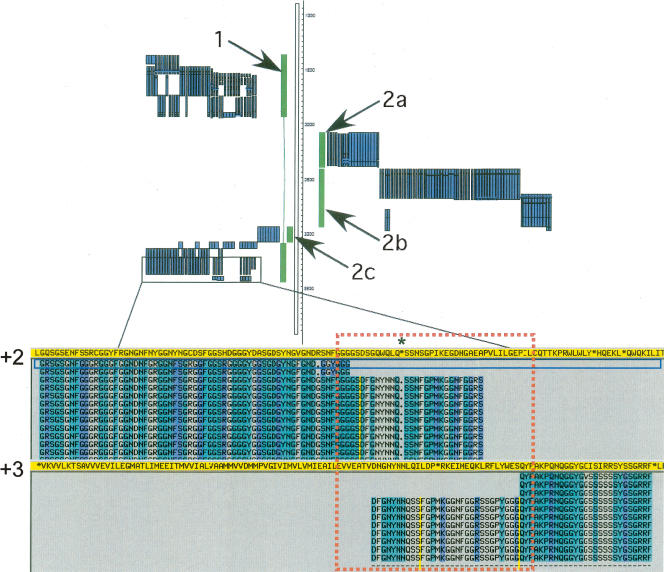
Figure 9.
Complexity in pseudogene annotation—insertion of one pseudogene into another. A set of “nested” pseudogenes (in green) was found in the ENm001 region with protein homology (shown in blue) supporting the annotation. This arrangement appears to have been generated through the insertion of a heterogeneous nuclear ribonucleoprotein A1 (HNRPA1) processed pseudogene (1) into the genome on the negative strand. This was followed by a second insertion event in which a transcript originating from the mitochondrial genome was transposed into the HNRPA1 pseudogene sequence. Gene order and orientation suggest that this mitochondria-derived sequence has undergone further rearrangement, including deletions, to leave an NADH dehydrogenase 2 (MTND2) pseudogene (2a) and an NADH dehydrogenase 4 (MTND4) pseudogene (2b) on the positive strand and a cytochrome B (CYTB) pseudogene (2c) on the negative strand. A view of the protein alignment for the 5′-end of the HNRPA1 pseudogene (in yellow) is shown with an in-frame stop codon (indicated by *) and a shift from frame +2 to +3 (highlighted by the red box) clearly visible.