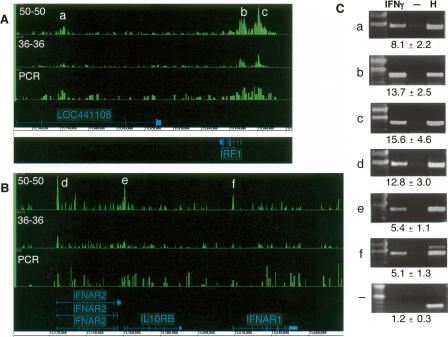
Figure 1.
Comparison of signal tracks. Signal enrichment tracks are plotted for the 50-b, 36-b, and PCR product array platforms for two different loci. Signals of STAT1 bound regions in IFNG-stimulated cells relative to untreated cells were derived from multiple biological replicates with one replicate hybridized per array (Methods; Supplemental Table 1). Annotations above the coordinate axis are for genes on the forward strand, and those below are for reverse-strand genes. Signal enrichment tracks are plotted to the same scale for the platforms displayed, from 0 to 2.5 in panel A and from 0 to 1.3 in panel B. (A) The IRF1 locus and flanking regions on Chromosome 5 (coordinates 131,770,000 to 131,870,000 from build NCBIv35 [hg17]). (B) The loci on Chromosome 21, which contain the cytokine receptors, IFNAR2, IL10RB, and IFNAR1 (coordinates 33,500,000 to 33,680,000). (C) Targets that have been validated by ChIP-PCR (shown) are indicated by symbols a through f. The lanes are labeled for ChIP DNA from IFNG-stimulated (IFNγ) cells, ChIP DNA from unstimulated cells, and for HeLa S3 genomic DNA. Fold enrichments, as calculated for several biological replicates (see Methods), are indicated for each target (a–f) and for a negative control region (−).