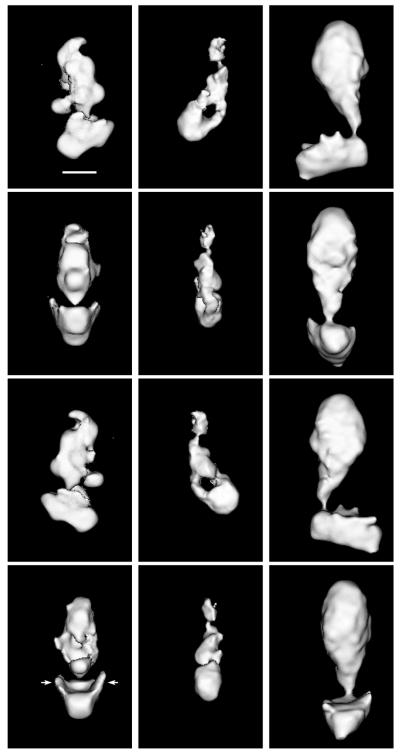
Figure 1.
Shown are 3-D structures of yeast Mediator (Left), murine Mediator (Center), and of the human TRAP complex (Right). The orientation of each complex in consecutive rows differs by 90°. (Top) Complexes in an orientation corresponding to the preferred orientation of the respective particles on the grid. (Bar = 100 Å.) The arrows near the head domain of the yeast Mediator indicate the “flaps” that must wrap around RNA polymerase II when the holoenzyme complex is formed. Cut-off levels for displaying the different maps were chosen to emphasize their overall domain organization. The resolution of the maps is 30–35 Å, as determined by the Fourier-ring correlation method (17), although a loss of resolution in the direction perpendicular to the plane of Top results from incomplete sampling of the structures in that direction.