Abstract
The effort of function annotation does not merely involve associating a gene with some structured vocabulary that describes action. Rather the details of the actions, the components of the actions, the larger context of the actions are important issues that are of direct relevance, because they help understand the biological system to which the gene/protein belongs. Currently Gene Ontology (GO) Consortium offers the most comprehensive sets of relationships to describe gene/protein activity. However, its choice to segregate gene ontology to subdomains of molecular function, biological process and cellular component is creating significant limitations in terms of future scope of use. If we are to understand biology in its total complexity, comprehensive ontologies in larger biological domains are essential. A vigorous discussion on this topic is necessary for the larger benefit of the biological community. I highlight this point because larger-bio-domain ontologies cannot be simply created by integrating subdomain ontologies. Relationships in larger bio-domain-ontologies are more complex due to larger size of the system and are therefore more labor intensive to create. The current limitations of GO will be a handicap in derivation of more complex relationships from the high throughput biology data.
Keywords: gene, ontology, function, annotation, vocabulary
Background
The protein function annotation problem is central to understanding of biological systems. A major challenge that is currently facing the biology community is to precisely define what action(s) constitutes a given function. The challenge lies in the fact that the same action is interpretable in different ways to different people if the context in which the action is taking place is not known. For example in proteins, the action “bind” describes association between two proteins. But left to itself, the action is open to further interpretation. For example: bind for catalysis, bind for transport or bind for regulation? Similar things can be said about actions like fusion, docking, porting, sensing, silencing and so on. It is apparent that some actions share parent child relationships and these natural relationships must be recognized for assisting us in the function annotation efforts.
Another degree of complexity in the definition of function comes from the description of functional levels. As is evident, function can be defined as any of a group of related actions contributing to a larger action in a living organism (Webster Dictionary definition). This means lower level functions will interact to generate higher-level function and at the same time, a lower level function will be part of many higher-level functions. These relationships among individual functions at a each level and among many levels must be established properly to identify a function and its order (order of a function is the number of component functions that makes it). The relationship between functions at different levels and also among themselves must also be identified.
Description
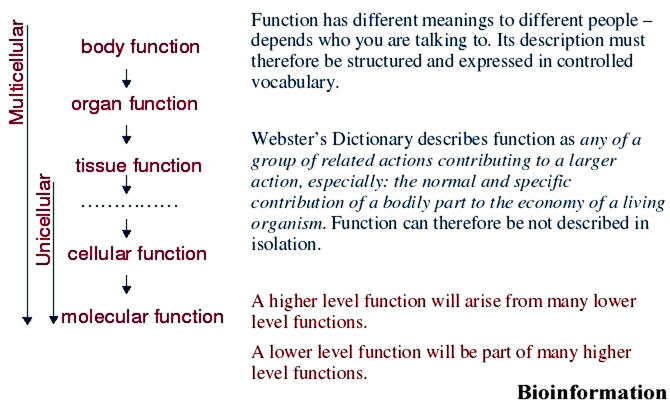
Gene Ontology (GO, http://www.geneontology.org) currently houses the most extensive set of description of molecular functions, biological processes and cellular components using controlled vocabulary. [1] Each description in these sets can be called a term, which are related to another by a directed edge. The graph that is thus formed is called a directed acyclic graph where in the detail of description improves as we go further away from the root term. The GO takes into account the dependencies of terms and parent child relationship. For example, in biological processes if either X biosynthesis or X catabolism exists, then the parent X metabolism is included. Similarly, if regulation of X exists, then the process X must also exist. However, all such logically related terms are not included in a defined manner. For example, in molecular function, binding activity is segregated from related activities like transport and catalysis. The current argument of GO is that binding terms should only be used in cases where a stable binding interaction occurs. It has been argued that if actions were indeed subdivided, splitting the catalysis of a reaction into steps such as “substrate binding”, “formation of unstable intermediate” or “attraction of electrons to positive charge” - will lead to saying that a reaction was actually a series of functions i.e., a process. There is certain paradox in the arguments here, because ontologies in GO are developed at three segregated levels: molecular function, biological process and cellular component. In reality, molecular functions are also processes involving atoms, and this fits nicely within our prescribed definition of function. There should naturally exist relationships that link molecular function to biological process, or any level that involves entities such as atoms, tissues, organs and so on (Figure 1).
Figure 1.
A proposed outline of hierarchy in an unified biological ontology describing function. Each species may have different organization of the hierarchies depending on its structural organization. While for unicellular organisms the hierarchy is restricted to cell; in multicellular organism this hierarchy can extend to tissues, glands, bones, organs, appendage, body and so on. In all these cases, the functional relationships must conform to the definitions proposed on the right panel. In our scheme, although we have restricted our hierarchy to molecular function, in principle it can be extended to molecular subdomains, atoms and electrons. The graph formed from the relationships is expected to be directed and acyclic to make it amenable to computations.
Therefore a critical challenge remains in understanding function terms in their proper perspective so that all functions at different levels can be related to each other. Certain effort is already ongoing [2 ], how we can develop Gene Ontology with clarity of logical relationships, which are also mathematically amenable to interpretation and useful for functional annotation. Open Biomedical Ontologies (OBO, http://obo.sourceforge.net/) are another effort in this direction. Although effort to build ontologies in a given domain is useful, its utility will be limited by lack of cross domain link, which cannot be overcome by simply integrating individual domain ontologies. Such larger bio-domain ontologies therefore need to be built almost from scratches, since the complexities of relationships are diverse, an essential requirement for understanding the complexity of biological actions. Therefore efforts to build ontologies in larger biological domains must be started so that the inherent biosystem complexities are truly resolved.
Because complexities of relations in large domains are manifold, only experts are properly qualified to define clear relations. With a worldwide community initiative, however, the possibilities of having these ontologies are not impractical. Voluntary contribution of these ontologies derived from experimental work of the authors, who are self-experts in the area, will benefit the effort. Research journals can encourage such contributions by making ontology submissions mandatory to a public database. It is arguably one of the most challenging frontiers facing the biology community today.
Footnotes
Citation:Pal, Bioinformation 1(3): 97-98 (2006)
References
- 1.Ashburner M, et al. Nature Genet. 2000;25:25. [Google Scholar]
- 2.SchulzeKremer S. In Silico Biol. 2002;2:179. [Google Scholar]