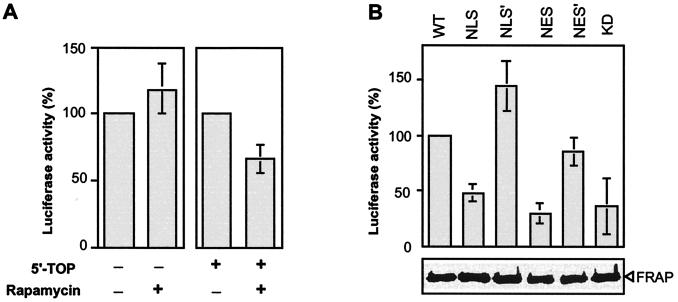
Figure 6.
Cytoplasmic–nuclear shuttling of FRAP regulates serum-stimulated rapamycin-sensitive translation. pCDNA3-luciferase cDNAs were transiently transfected into HEK293 cells. Transfected cells were serum-starved for 36 h, followed by serum stimulation for 3 h before cell lysis. Luciferase assays were carried out by using the luciferase assay system (Promega) according to the manufacturer's manual. Activities shown here correspond to luciferase expression during the 3-h serum stimulation. (A) Rapamycin (100 nM) was added during the 3-h serum stimulation of cells transfected with luciferase cDNA with or without eEF2 5′ untranslated region containing the 5′-TOP. The data shown are for activities relative to that in untreated cells. (B) Cotransfection of 5′-TOP-luciferase with variously tagged Myc-FRAPs was followed by luciferase assays performed after serum stimulation for 3 h in the presence of 100 nM rapamycin. Designation of various FRAP proteins is described in the legend to Fig. 4. All FRAP constructs, including wild type, contained the S2035T mutation. The data shown are for activities relative to that for wild type. FRAP protein expression was monitored by Western analysis.