Figure 2.
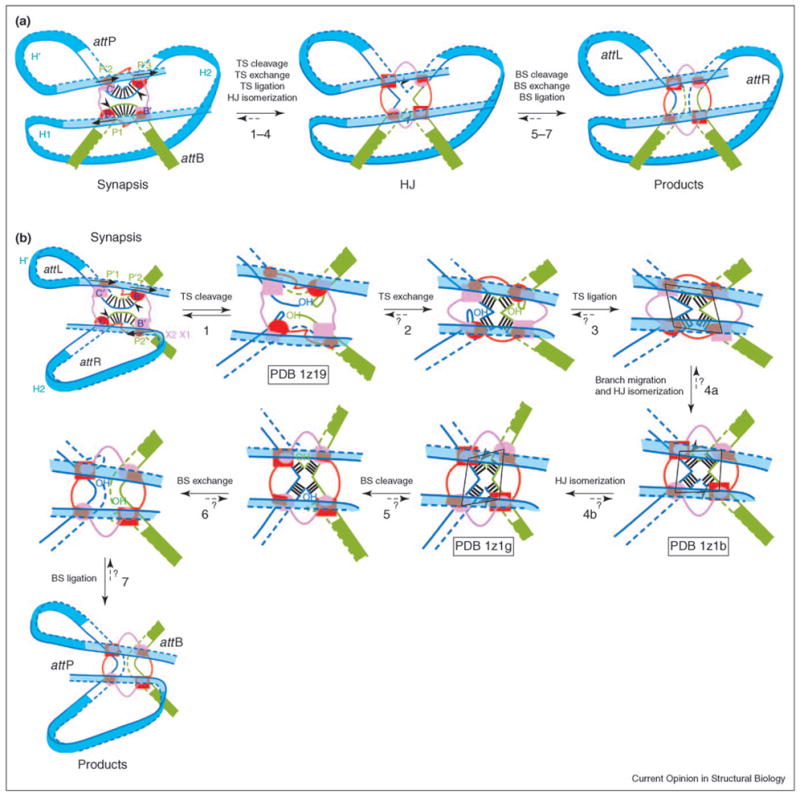
Reaction steps for (a) integration and (b) excision. Reaction steps are illustrated using the current model of the complete higher order recombination complex. Top strands (TS, continuous lines) are cleaved first and bottom strands (BS, dashed lines) are cleaved second to resolve the HJ. Cleavage sites (black arrowheads) are located on either side of the seven base pairs of the overlap region [vertical black lines in the cartoons for synapsis (a,b) and steps 3–6]. Active and inactive Int monomers are colored red and pink, respectively. Top-strand-cleaving Ints bound to core sites C and B, and bottom-strand-cleaving Ints bound to C′ and B′ are drawn as ovals and rectangles, respectively. The smaller ovals and rectangles represent the N-domains, which are connected to the C-terminal domains (bigger rectangles and ovals) by a flexible linker (red and pink lines). Note that the shapes of the arm DNA loops are not drawn for the reaction intermediates in (b). We do not know which of the steps (maybe all of them) are kinetically irreversible, as illustrated by short dashed arrows and question marks. The shape of the Int tetramer (shown as a black parallelogram in steps 3, 4a and 4b) changes during HJ isomerization, and Int activity switches from top strand to bottom strand cleavage. Steps 1, 4a and 4b are represented by crystal structure PDB codes 1z19, 1z1b and 1z1g [C75 Int (truncated Int 75–356) tetramer bound to core DNA; a post-strand-exchange complex containing an Int tetramer, two core DNA segments and two short arm DNAs; and an Int tetramer bound to a HJ and two short arm DNA segments], respectively [8••]. The more pronounced twofold symmetry of the second structure (step 4b) suggests that it has undergone further HJ isomerization than the structure in step 4a. During HJ isomerization, the HJ also performs branch migration of one base pair (step 4a). Consequently, the two bottom-strand-cleaving Ints end up being separated by six base pairs, as opposed to the eight base pair separation between the top-strand-cleaving Ints. This movement is thought to rearrange catalytic domain interactions in the Int tetramer, thus activating the bottom-strand-cleaving Ints. The structures suggest that arm DNA binding and the conformation of the N-domain tetramer favor the HJ conformation that activates the bottom-strand-cleaving Ints.
