Figure 1.
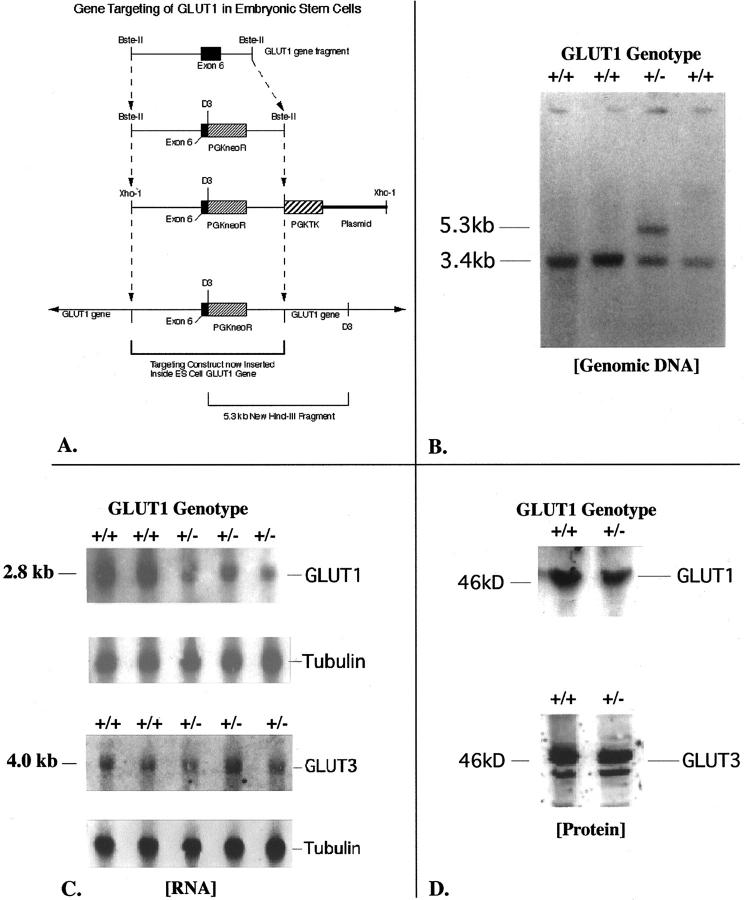
A: This diagram depicts the restriction map and modifications made to a cloned GLUT1 mouse genomic DNA fragment containing exon 6, to build the final targeting (homologous recombination) construct shown. The original fragment was 12.2 kb long. A 0.8-kb HindIII fragment including the 3′ end of exon 6 was removed, and the PGKneoR cassette inserted here in the 3′ to 5′ orientation. The pGKTK cassette was ligated to the 3′ end of the genomic fragment. The finished product was carried in pBS SKII+. This circular DNA was linearized at a unique 5′ XhoI site to produce the final homologous recombination construct as shown. The plasmid flanks the pGKTK gene at the 3′ end of the linearized construct. B: This Southern blot of HindIII-digested genomic DNA from four ES cell clones was hybridized with an adjacent 1.66-kb GLUT1 cDNA probe that would recognize both the normal gene at 3.4 kb and the targeted gene at 5.3 kb. The clone in lane 3 demonstrates the expected genotype for successful gene targeting at the GLUT1 locus. The homologous recombination construct inserts into one copy of the GLUT1 gene. The HindIII restriction enzyme cuts both inside the construct, and beyond the 3′ end of the construct in adjacent ES cell genomic GLUT1 DNA. This allows the GLUT1 probe to recognize the same HindIII band (at 5.3 kb) as a PGKneoR probe when the homologous recombination construct containing the PGKneoR sequence has successfully inserted into the ES cell GLUT1 gene. C: Northern blots of GLUT1 and GLUT3 mRNAs in ES cells are shown here. The GT1(+/+) ES cells are the nontargeted cells, whereas the GT1(+/−) ES cells are the heterozygous GLUT1-targeted ES cells. The β-tubulin transcript is shown for the same lanes to confirm even lane loading and normalize GLUT1 transcript levels to the β-tubulin housekeeping mRNA level. Optical scanning densitometry was used to determine the 49 ± 4% decrease of GLUT1 mRNA with heterozygous targeting of the GLUT1 gene. In contrast, no significant change in GLUT3 mRNA was detected in these targeted ES cells. D: GLUT1 protein in ES cells was detected as a band at ∼46 kd, and this band competed away with 25 μg/ml of the specific GLUT1 peptide antigen. GLUT3 protein was identified at 44 to 46 kd, possibly because of variations in glycosylation, and these bands competed away with 25 μg/ml of specific GLUT3 peptide antigen. GT1(+/+) ES cells are the nontargeted ES cells, and GT1(+/−) are the heterozygous GLUT1-targeted ES cells.