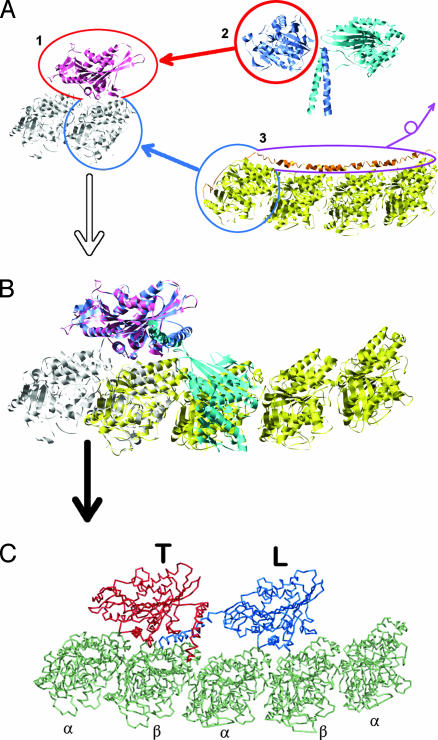
Fig. 2.
Procedure to construct the two-headed kinesin/MT-protofilament model. (A) Three structures from the PDB are used. 1, Single-headed kinesin (KIF1A) bound on tubulin (PDB ID 1IA0); 2, two-headed kinesin (PDB ID 3KIN); 3, two consecutive tubulin complexed to the stathmin-like domain (PDB ID 1FFX). (B) We overlapped chain A (blue) of 3KIN onto chain K of 1IA0, and we overlapped chains A and B of 1FFX (α-domain) onto chain B of 1IA0, which leads to the structure shown. The structural homology (Cα backbone rmsd = 1.6 Å) between KIF1A and a head of the two-headed kinesin are sufficient that one of the kinesin heads fits to the tubulin binding site. Although the sequence difference between KIF1A and the conventional kinesin (sequence identity, ≈45%) may affect the strength of interactions between kinesin and tubulin, leading to a different binding affinity of conventional kinesin from KIF1A, we assume that the binding orientation of the two-headed kinesin is similar to that of KIF1A on the tubulin. After the structure overlap, chains C and D of 3KIN are internally rotated around a few positions in the neck-linker (324–338) until chains C and D are placed in the vicinity of the binding site of tubulin, which is designed to be identical to the interface between the kinesin T and the tubulin. (C) We performed the simulation to relax the kinesin structure on the MT and obtained the structure shown.