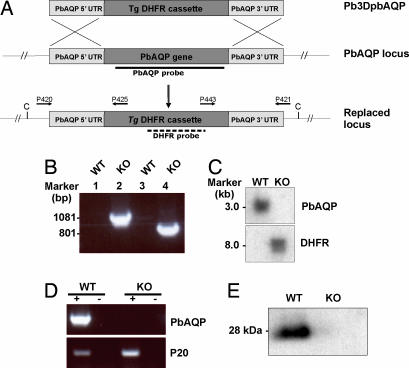
Fig. 4.
Molecular characterization of PbAQP-null parasites. (A) Schematic representation of the PbAQP WT locus, the targeting construct (pB3DPbAQP) used to delete the endogenous PbAQP locus, and the PbAQP locus after integration. The locations of primers used in PCRs to confirm integration are indicated. The location of the PbAQP and DHFR probes and the expected size of the fragments detected by Southern blot analysis of genomic DNA digested with ClaI (C) are shown. (B) PCR analysis of genomic DNA to confirm the integration of pB3DPbAQP into the endogenous PbAQP locus by using the primers shown in Fig. 4A. The PCR-amplified products were run on 1% gel (wt/vol) agarose/TAE gel and stained with ethidium bromide. Lanes 1 and 2, template DNA with primers P420/P425; lanes 3 and 4, template DNA with primers P443/P421; lanes 1 and 3, WT parasite; lanes 2 and 4, PbAQP-null parasite (KO). The molecular sizes are indicated (in kilobases). (C) Southern blot analysis. Genomic DNA from WT and KO parasites was digested with ClaI and probed with PbAQP or DHFR probes. The molecular sizes are indicated (in kilobases). (D) RT-PCR. Total RNA was extracted from WT and KO parasites and reverse transcribed (+ lanes) by using primers P403/P404 and P401/P402. Lanes marked with (−) show results without reverse-transcriptase step. (E) Western blot analysis of protein lysates from WT and PbAQP-null parasites probed with PbAQP antibodies.