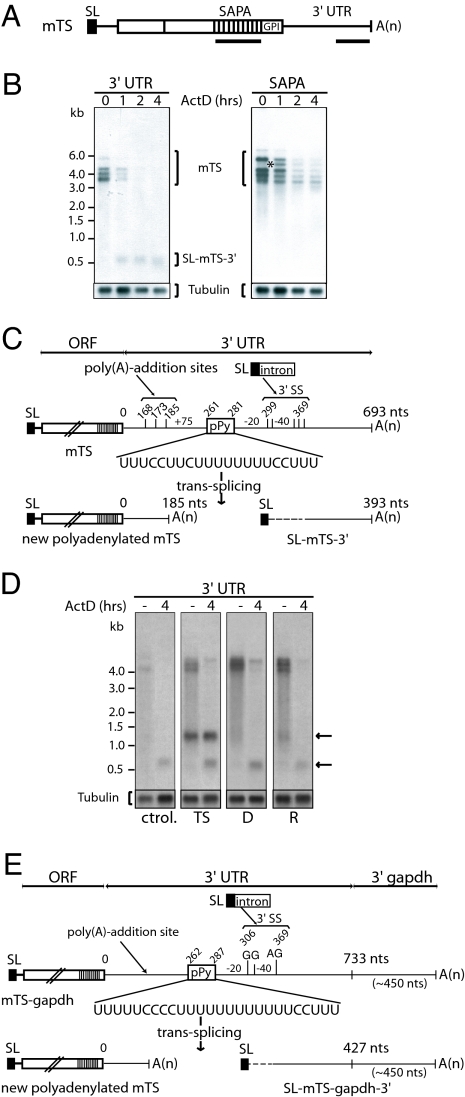
Fig. 3.
Trans-splicing/polyadenylation events in the 3′ UTR of TS RNAs. (A) Scheme showing the location of the probes used in these experiments. (B) Northern blots of poly(A)+ RNA from parasites treated with ActD for the indicated times. Hybridizations were performed with mTS-3′ UTR and SAPA probes. The asterisk indicates the detection of a new band after 1 h of ActD treatment. A β-tubulin probe was used for loading control. (C) Structure of mTS, the new polyadenylated mTS, and the SL-mTS-3′ RNAs generated by trans-splicing. Nucleotides where trans-splicing and polyadenylation events take place are indicated. 3′SS, 3′ trans-splicing sites; pPy, polypyrimidine tract. The drawings are not in scale. One representative sequence is shown. (D) Northern blots of total RNA from wild-type parasites (ctrol.) and parasites transfected with different pTEX constructs (D, TS with a 90-nt deletion including the pPy sequence and two 3′SS, R, TS in which the above-mentioned 90-nt fragment has been replaced by another sequence) without treatment (−) or incubated with ActD for 4 h (4). Hybridizations were performed with mTS-3′ UTR probe. The upper arrow indicates the SL-mTS-gapdh-3′ species. The lower arrow shows the endogenous SL-mTS-3′. (E) Structure of the recombinant mTS-gapdh mRNA and the SL-mTS-gapdh-3′ RNA generated by trans-splicing. 3′SS, acceptor splice site; pPy, polypyrimidine tract.