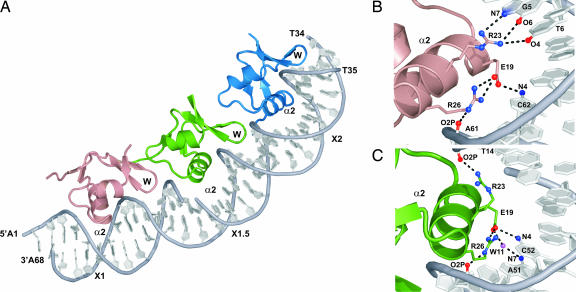
Fig. 3.
The structural basis of cooperative binding is revealed by crystallography. (A) X-ray crystal structure of Xis bound to the Xis binding region. Xis monomers bound to the X1, X1.5, and X2 sites are colored dark salmon, green, and blue, respectively. The nick present in the top strand of the DNA duplex is a result of annealing three complementary DNA strands (Fig. 1B). The proteins adopt nearly identical “winged” helix structures (the backbone coordinates of residues Tyr-2–Val-50 in each protomer can be superimposed with a rmsd of 0.22 Å). The secondary structural elements in this fold are arranged in the following configuration: β1-α1-L-α2-B1-β2-β3-W-β4-B2-β5, where L, W, and B are the loop, wing, and bulge structures, respectively. Residues within each secondary structural element are as follows: β1 (Tyr-2–Thr-4), α1 (Leu-5–Arg-11), L (Gln-12–Ser-17), α2 (Leu-18–Arg-26), B1 (Glu-27–Arg-29), β2 (Ile-30–Phe-31), β3 (Val-35–Asp-37), W (Gly-38–Arg-39), β4 (Glu-40–His-44), B2 (Glu-45–Ala-47), and β5 (Val-48–Lys-49). (B and C) Residues within Xis–DNA interfaces involved in hydrogen bonding. (B) Shown is the interface between DNA and the Xis monomer bound at X1. (C) Shown is the interface between DNA and the Xis monomer bound at X1.5.