TABLE 1.
Data set descriptions and summary results
| Data set | MAF classa | Populationsb | Individuals | Loci |
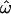 %c %c
|
CC%d | CT%e | CT%, single locusf |
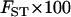 g g
|
% rare and commonh | % fixed and commoni |
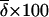 j j
|
% het.k |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Insertions | All | Distinct (3) | 219 | 175 | 10 | 0.46 | 0 | 56 | 14 | 21 | 10 | 18 | 32 |
| Insertions | Rare | Distinct (3) | 219 | 21 | 47 | 58 | 39 | 66 | 12 | 57 | 33 | 5.7 | 7.0 |
| Insertions | Common | Distinct (3) | 219 | 154 | 9.1 | 0.46 | 0 | 55 | 14 | 16 | 7.1 | 20 | 36 |
| Insertions | All | All (4) | 259 | 175 | 15 | 3.1 | 1.2 | 65 | 12 | 23 | 11 | 15 | 32 |
| Insertions | Rare | All (4) | 259 | 21 | 50 | 76 | 46 | 73 | 8.3 | 62 | 43 | 4.3 | 7.5 |
| Insertions | Common | All (4) | 259 | 154 | 13 | 3.1 | 0.77 | 64 | 12 | 18 | 6.5 | 17 | 36 |
| Microarray | All | Distinct (3) | 137 | 9,922 | 0 | 0 | 0 | 56 | 15 | 26 | 12 | 20 | 32 |
| Microarray | Rare | Distinct (3) | 137 | 920 | 17 | 7.3 | 0 | 65 | 10 | 87 | 56 | 9.5 | 10 |
| Microarray | Common | Distinct (3) | 137 | 9,002 | 0 | 0 | 0 | 55 | 15 | 19 | 7.8 | 21 | 34 |
| Microarray | All | All (8) | 278 | 9,922 | 3.1 | 10 | 1.1 | 82 | 13 | 44 | 27 | 18 | 32 |
| Microarray | Rare | All (8) | 278 | 851 | 36 | 72 | 1.4 | 84 | 8.9 | 98 | 88 | 7.9 | 11 |
| Microarray | Common | All (8) | 278 | 9,071 | 3.3 | 8.3 | 1.1 | 81 | 13 | 39 | 21 | 19 | 34 |
| Resequenced | All | All (3) | 209 | 14,233 | 8.2 | 0 | 0 | 57 | 13 | 40 | 29 | 11 | 17 |
| Resequenced | Rare | All (3) | 209 | 8,389 | 29 | 45 | 0.96 | 59 | 9.6 | 51 | 43 | 4.6 | 4.7 |
| Resequenced | Common | All (3) | 209 | 5,844 | 5.9 | 0.96 | 0 | 53 | 14 | 23 | 8.4 | 19 | 34 |
Sets of loci with MAF < 0.1 (rare), MAF > 0.1 (common), or any MAF (all).
Sets of individuals representing all populations in a data set or only the more distinct populations.
Dissimilarity fraction.
Centroid misclassification rate.
Population trait value misclassification rate.
Trait value misclassification rate based on a single locus, averaged over all loci.
The proportion of variance in allelic frequencies attributable to population differences.
Percentage of loci with MAF < 0.1 in one population and MAF > 0.1 in another.
Percentage of loci that are monomorphic in one population and polymorphic with MAF > 0.1 in another.
Average difference in allele frequency between populations, averaged across alleles, loci, and population pairs.
Percentage of individuals heterozygous at each locus, averaged across all loci.
