TABLE 2.
Comparison of analysis methods
| Estimation method | U |  |
β |
|---|---|---|---|
| Data-driven estimates of the distribution of fixed mutations | |||
Detected mutationsa (Umin, 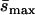 ) ) |
0.011 | 0.169 | 0.076 |
| MLEb (nDEL = 56) | 0.028 | 0.066 | 0.162 |
| MLEb (nDEL ≤ 56) | 0.017 | 0.109 | 0.116 |
| Data-driven estimates of the distribution of new mutationsc | |||
| MLE (nDEL = 56) | 0.030 | 0.093 | 0.204 |
| MLE (nDEL ≤ 56) | 0.021 | 0.142 | 0.141 |
| Model-based estimates of the distribution of fixed mutations | |||
| Bateman–Mukai | 0.035 | 0.0269 | 0d |
| Maximum likelihoode | 0.045 | 0.0207 | 0 |
Mutations whose fitness effects were detected by the stepwise regression analysis.
Estimated using the method of Otto and Jones (2000) by specifying that all of the accumulated mutations were deleterious (nDEL = 56) or allowing a fraction of the accumulated mutations to be neutral (nDEL ≤ 56).
Estimated by correcting the estimated fixed mutation distributions for the probability that mutations of various effect sizes were lost due to selection during plaque growth.
The Bateman–Mukai analysis method does not estimate Var(s). Rather, it assumes Var(s) = 0.
Estimated using the method of Keightley (1994).
