TABLE 1.
Limits to multivariate trait variation
| Sample size
|
||
|---|---|---|
| Eigenvalue rank | 75 | 300 |
| 1 | 6.41e-1 ± 3.20e-2 | 6.22e-1 ± 2.80e-2 |
| 2 | 3.09e-1 ± 1.86e-2 | 3.08e-1 ± 1.74e-2 |
| 3 | 1.56e-1 ± 1.25e-2 | 1.56e-1 ± 1.17e-2 |
| 4 | 7.37e-2 ± 7.32e-3 | 7.67e-2 ± 7.22e-3 |
| 5 | 3.63e-2 ± 4.69e-3 | 3.87e-2 ± 4.62e-3 |
| 6 | 1.72e-2 ± 2.71e-3 | 1.92e-2 ± 2.78e-3 |
| 7 | 7.85e-3 ± 1.60e-3 | 9.48e-3 ± 1.69e-3 |
| 8 | 3.31e-3 ± 8.07e-4 | 4.38e-3 ± 9.05e-4 |
| 9 | 1.27e-3 ± 4.42e-4 | 1.91e-3 ± 5.04e-4 |
| 10 | 5.23e-4 ± 2.44e-4 | 8.12e-4 ± 2.95e-4 |
| 11 | 2.02e-4 ± 1.34e-4 | 3.33e-4 ± 1.74e-4 |
| 12 | 9.08e-6 ± 1.78e-5 | 1.27e-4 ± 9.90e-5 |
| 13 | 0.0 | 6.02e-5 ± 5.00e-5 |
| 14 | 0.0 | 1.49e-5 ± 1.60e-5 |
| 15 | 0.0 | 1.19e-6 ± 2.34e-6 |
| 16 | 0.0 | 0.0 |
When the effective population size or genomic mutation rate for a set of characters is low enough, some principal components are expected to have zero variance associated with them. For instance, the eigenvalues are given ranked in order from largest to smallest when 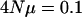 per locus and the number of loci is 10. The M matrix had dimensionality of 25 with the same variance for each principal component.
per locus and the number of loci is 10. The M matrix had dimensionality of 25 with the same variance for each principal component.
