Figure 1.
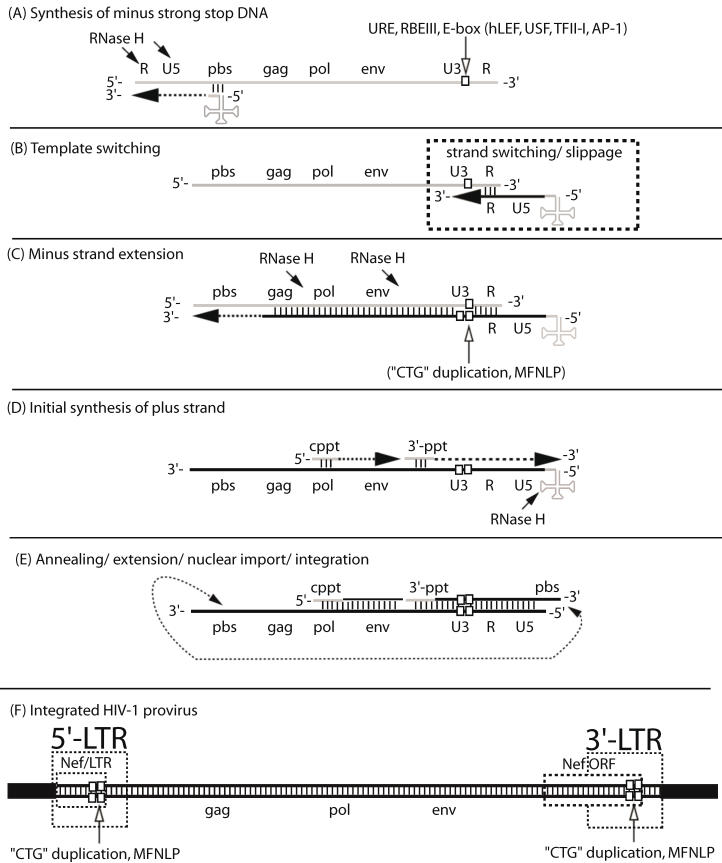
The MFNLP is generated during reverse transcription. (A) Synthesis of minus strong stop DNA, begins with t-RNA (clover-leaf structure) binding to the primer binding site (pbs), followed by RNA-templated polymerization of minus strong stop DNA (dashed arrow). RNase H degrades the template RNA after polymerization (arrows); (B) Template switching to the other RNA genome copy or to the same RNA, occurs via complementary sequences in the Repeat region (R). During the template switch, slippage can occur; (C) Minus strand extension could combine with slippage, templated and non-templated polymerization, resulting in duplication of the URE/RBEIII/partial E-box region (single white box, in RNA U3 region), generating a first copy of the MFNLP (two white boxes in U3 region), in the minus DNA. RNAse H continues to degrade the RNA genome, leaving just the polypurine tracts. (D) Initial synthesis of plus strand DNA begins from the central polypurine tract (cppt) and from the 3'-polypurine tract (3'-ppt), resulting in synthesis of a plus strand MFNLP (four white boxes); (E) Annealing of the free pbs, further extension and rearrangements, are followed by nuclear import and integration of the proviral HIV-1 sequences. (F) The final integrated proviral sequences contain a 5'-LTR and a 3'-LTR with U3, R, U5 sequences. In this model, both the 5' and 3'-LTR would contain the MFNLP. Note the Nef ORF in the 3'-LTR and the Nef/LTR sequences in the 5'-LTR. Integrity of the Nef TGA stop codon (ACTGCTGA) is critical for RBF-2 binding to the MFNLP.