Figure 1.—
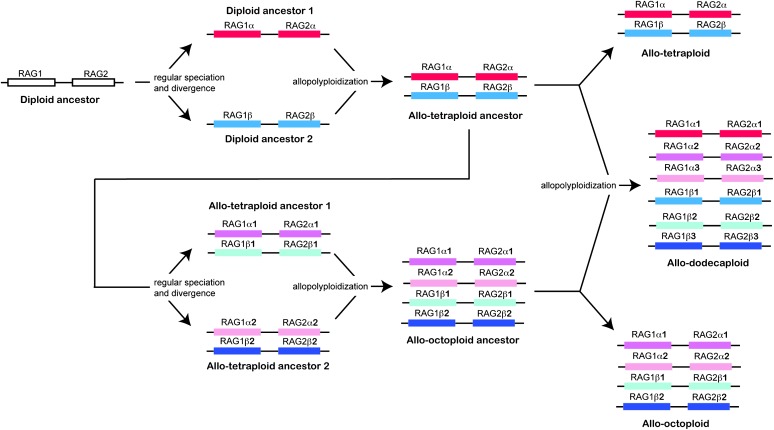
Diploidization of allopolyploid genomes means that recombination between alleles of different paralogs of the same gene is rare. The evolutionary history of linked paralogs in diploidized allopolyploid genomes, therefore, can be inferred by combining information on synteny with information on the evolutionary relationships among paralogs of each gene. The RAG1 and RAG2 genes, for example, are tightly linked. The evolution of the expected number of paralogs of each of these genes in species with different ploidy levels (tetraploid, octoploid, and dodecaploid) if no deletion or degeneration occurred is depicted. An allotetraploid species inherits a linked set of RAG1 and RAG2 α paralogs from diploid ancestor 1 and a linked set of RAG1 and RAG2 β paralogs from diploid ancestor 2. An allo-octoploid species inherits two sets of linked RAG1 and RAG2 paralogs from two different allotetraploid ancestors. In these species the α1 and α2 paralogs of RAG1 and RAG2 are derived from diploid ancestor 1 and the β1 and β2 paralogs of RAG1 and RAG2 are derived from diploid ancestor 2. An allododecaploid inherits an allo-octoploid and an allotetraploid genome and has three α paralogs (α1, α2, α3) derived from diploid ancestor 1 and three β paralogs (β1, β2, β3) derived from diploid ancestor 2. Note that the observed number of functional RAG1 and RAG2 paralogs is less than this expectation in most (RAG1) or all (RAG2) species of Xenopus due to degeneration or deletion.