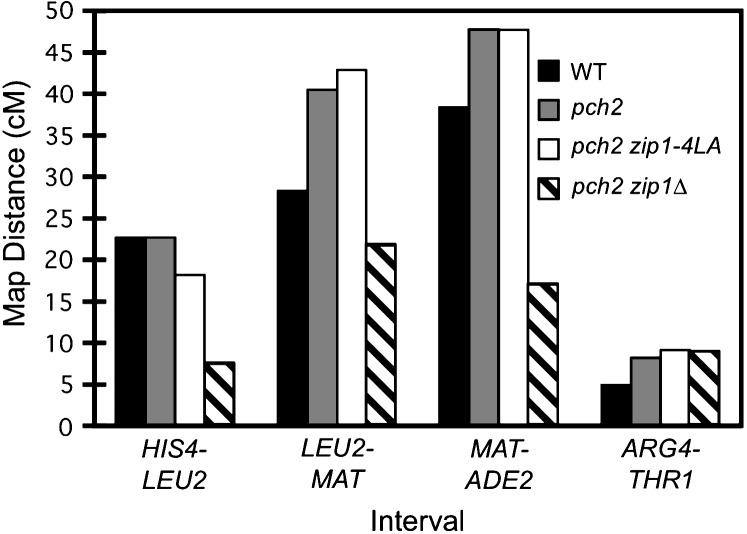
Figure 7.—
Tetrad analysis of pch2 zip1-4LA strains. Shown is the map distance (centimorgans) in three intervals on chromosome III and in one interval on chromosome VIII for wild-type (NMY650), pch2 (NMY654), pch2 zip1-4LA (NMY661), and pch2 zip1Δ (NMY664) strains in the BR1919-8B diploid background. Using the G-test of homogeneity, there is no statistically significant difference between map distances in pch2 and pch2 zip1-4LA for any interval: P = 0.460 for HIS4-LEU2; P = 0.617 for LEU2-MAT; P = 0.843 for MAT-ADE2; P = 0.884 for ARG4-THR1.