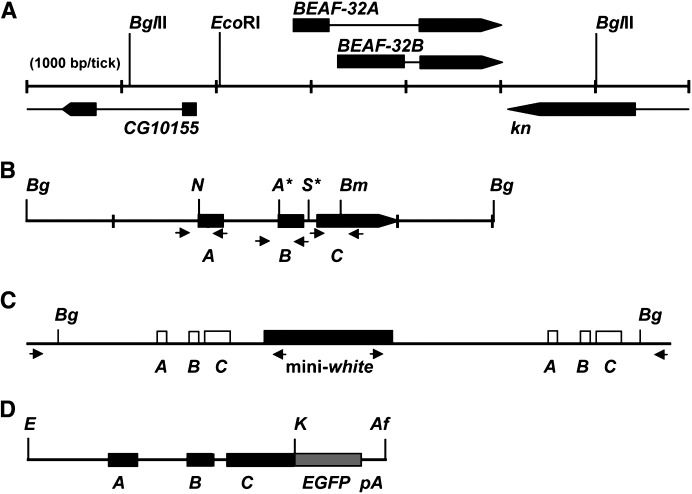
Figure 1.—
Strategy for targeted mutagenesis by homologous recombination. (A) Map of the BEAF gene, showing part of the upstream divergent CG10155 gene and downstream convergent knot (kn) gene. Arrows indicate the direction of transcription, and thin lines represent introns. Note the unique 5′ exons and the shared 3′ exon for BEAF-32A and BEAF-32B. (B) The BEAF gene was cloned as a 4.9-kb BglII fragment (gBF). Mutations were introduced at four locations to make the mutant mBF clone. The mBF gene was used for targeted mutagenesis by ends-in homologous recombination (Rong et al. 2002). Bg, BglII sites; N, destroyed NsiI site; A*, created ApaI site; S*, created I-SceI site; Bm, destroyed BamHI site. A, coding sequences unique to 32A; B, coding sequences unique to 32B. C, coding sequences common to both 32A and 32B. Arrows indicate primer pairs used for PCR. See materials and methods for details. (C) Schematic of the gene duplication expected from ends-in homologous recombination, with the mini-white marker gene between the duplicated BEAF gene. Arrows indicate primer pairs used for gene-specific PCR amplification of the 5′ or 3′ gene copy. (D) Schematic of the GFBF gene. The stop codon of the BEAF gene was converted to a KpnI site. Genomic BEAF sequences on an EcoRI–KpnI fragment were inserted upstream of EGFP sequences in the correct reading frame, with an SV40 polyadenylation sequence downstream of the EGFP sequences. E, EcoRI; K, KpnI; Af, AflII. See materials and methods for details.