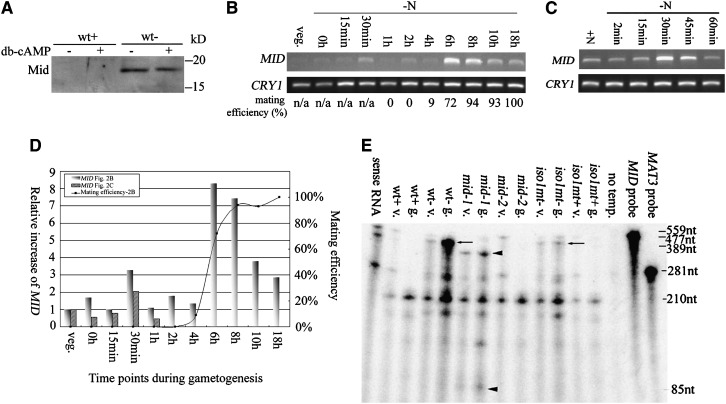
Figure 2.—
Patterns of MID expression. (A) Western blot of Mid in wild-type plus (wt+) and minus (wt−) gametes with or without the addition of db-cAMP. (B) RT–PCR of MID during gametogenesis. Synchronous wild-type minus cells were transferred to nitrogen-free (−N) media and samples were collected at various time points as indicated. RT–PCR products of poly(A) selected RNA were detected by ethidium bromide staining. CRY1, encoding ribosomal protein S14, is used as an internal control. Mating efficiencies of cells when samples were collected are standardized using mating efficiencies of wild-type tester cells. (C) RT–PCR of MID during early gametogenesis. Wild-type minus cells from 3-day-old TAP plates were transferred to nitrogen-free media and collected at various time points as indicated. Mating efficiencies of cells were not determined since vegetative cells do not differentiate into gametes within 1 hr. (D) Relative increases of MID during gametogenesis. The expression levels of MID in B and C were obtained by quantitation of the MID RT–PCR signals with the internal loading control, CRY1, and standardized by the relative amount of MID in vegetative cells. The relative increases of MID were plotted against time points when samples were removed during gametogenesis. The mating efficiency of individual samples from B is also plotted. (E) Expression of MID in vegetative cells. Total RNA isolated from wild-type and various mutant cells was hybridized with both a MID antisense RNA probe and a MAT3 antisense RNA probe and subjected to the ribonuclease protection assay. Arrows and arrowheads indicate the protected fragment of MID. mid-1, MID mutant with point mutations; mid-2, MID deletion mutant; iso1 mt−, an isoagglutination mutant; and iso1 mt+, mutant that carries the same mutation as in iso1 mt− but has a normal plus phenotype. The MID probe, 559 nucleotides (nt); the protected MID messages in wild-type minus and iso1 mt− cells, 477 nt (arrows); and the protected MID fragments in mid-1, 389 and 85 nt, respectively (arrowheads). MAT3, encoding a retinoblastoma homolog (Umen and Goodenough 2001), was used as an internal loading control. The MAT3 probe, 281 nt; and the protected MAT3 message, 210 nt. The third-to-last lane, no template, serves as a negative control with no RNA template.