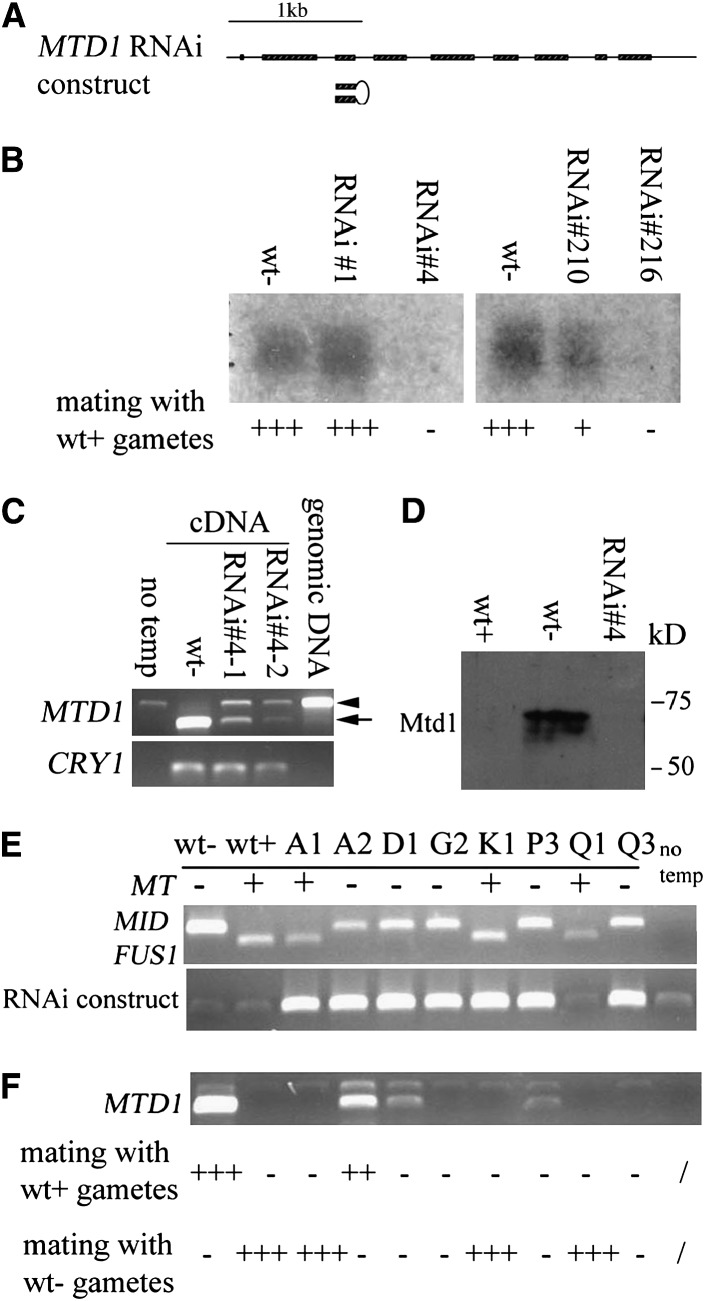
Figure 5.—
RNAi of MTD1 in minus cells. (A) Structure of the MTD1 gene and RNAi construct. Boxes, exons; lines, 5′-UTR, introns, and 3′-UTR. RNAi hairpin structure is presented by inverted pairs of exon 3 with intron 3 serving as the loop. The construct was driven by a constitutive HSP70A/rbcS2 promoter. (B) Northern blotting of MTD1 levels in different RNAi lines. The mating abilities of individual lines with wild-type plus gametes are indicated: +++, strong mating efficiency (80–100%); +, weak mating efficiency (20–50%); −, no mating. (C) RT–PCR of MTD1 in strain 4. Poly(A) RNA was isolated from two different cultures (#4-1 and #4-2). The arrow indicates the amplified cDNA fragment. The arrow head represents a weak contaminating genomic DNA fragment amplified in PCR. CRY1 is used as an internal control. The “no temp” control served as a negative control for PCR with no addition of DNA template. “Genomic DNA” served as a positive control for PCR by using genomic DNA as template. (D) Western blotting of Mtd1 in wild type and in the Mtd knockdown strain 4. (E) PCR of genomic DNA from progeny obtained from a cross between wt+ and strain 4. (Top) PCR of MID and FUS1 to determine mating types of individual progeny. (Bottom) PCR to detect the existence of RNAi construct. “No temp,” no DNA template control. (F) RT–PCR to detect the expression level of MTD1 in these progeny. The mating efficiency of individual progeny with either wild-type plus or wild-type minus gametes is indicated: +++, strong mating efficiency (80–100%); ++, moderate mating efficiency (50–80%); +, weak mating efficiency (20–50%); −, no mating.