TABLE 1.
Likelihood-ratio tests and model selection results
|
P-values for likelihood-ratio test of: |
||||||
|---|---|---|---|---|---|---|
| Strain |
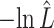 SS (P-value absolute g.o.f.) SS (P-value absolute g.o.f.) |
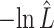 HT (P-value absolute g.o.f.) HT (P-value absolute g.o.f.) |
 VS VS |
SS vs. HT | SS vs. VS | HT vs. VS |
| H2 | NA | 1 | NA | NA | ||
| R28 | 49.77485 (0.1304) | 49.77485 (0.2457) | NA | 1 | NA | NA |
| P21 | 45.738 (0.00052) | 30.24335 (0.5313) | NA | 1.86534E-07 | NA | NA |
| P18 | 246.0254 (0) | 245.2713 (0) | 104.4029 | NA | 4.20413 × 10−61 | 3.14139 × 10−63 |
| S34 | 189.7365 (0) | 175.8767 (0) | 64.91768 | NA | 7.8379 × 10−54 | 3.45158 × 10−50 |
| S96 | 644.6472 (0) | 520.86 (0) | 59.69714 | NA | 2.4877 × 10−253 | 1.3758 × 10−202 |
| S55 | 227.3623 (0) | 227.3623 (0) | 83.82895 | NA | 6.26173 × 10−62 | 2.16624 × 10−64 |
The estimated negative log-likelihood score 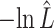 for each model (SS, segregation selection; HT, horizontal transfer; VS, variable selection) and strain combination is given under the first three columns. The better the model fit, the lower its computed
for each model (SS, segregation selection; HT, horizontal transfer; VS, variable selection) and strain combination is given under the first three columns. The better the model fit, the lower its computed 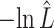 value. For the SS and HT model columns, the absolute goodness-of-fit (g.o.f.) P-values in parentheses indicate whether the model describes adequately (P-value > 0.05) or not (P-value < 0.05) the data at hand. Finally, the last three columns present the P-values for the likelihood-ratio tests to evaluate which model was the best for describing the data.
value. For the SS and HT model columns, the absolute goodness-of-fit (g.o.f.) P-values in parentheses indicate whether the model describes adequately (P-value > 0.05) or not (P-value < 0.05) the data at hand. Finally, the last three columns present the P-values for the likelihood-ratio tests to evaluate which model was the best for describing the data.
