Abstract
Aims
The objective of this study was to evaluate the potential uses of relative abundance, relative activity approaches and inhibitory monoclonal antibodies (mAbs) in the characterization of CYP enzymology in early drug discovery.
Methods
Intrinsic clearance estimates for the oxidation of ethoxyresorufin (a selective probe of CYP1A2 activity), tolbutamide (CYP2C9), S-mephenytoin (CYPC19), dextromethorphan (CYP2D6) and testosterone (CYP3A4) were used to determine relative activity factors (RAFs). CLint values were determined for the metabolism of 14 drugs in human liver microsomes (HLM) and for these major CYPs. The relative contribution of each individual CYP to the oxidation of each drug was then assessed using relative abundance and activity techniques in addition to inhibitory mAbs.
Results
Relative abundance and activity methods as well as inhibitory mAbs qualitatively assigned the same CYP isoform as predominantly responsible for the clearance of each drug by HLM. Metabolism catalysed by CYP1A2, 2C9, 2D6 and 3A4 was also predicted to be quantitatively similar using both abundance and activity techniques. However, the relative contribution of the polymorphic CYP2C19 appeared to be over-estimated approximately two-fold using recombinant CYP compared with that from the HLM and mAb approach.
Conclusions
All three methods investigated in this study appear suitable for use in the characterization of the CYP metabolism of new chemical entities produced during early drug discovery.
Keywords: HLM monoclonal antibody, intrinsic clearance, reaction phenotyping, recombinant CYP
Introduction
The cytochrome P450 (CYP) enzyme family catalyses the oxidation of a plethora of endogenous compounds (steroid hormones, eicosanoids, retinoids, vitamins) and xenobiotics (drugs, chemicals, pollutants) [1]. Interindividual variation in drug metabolism by CYPs may be associated with dramatic perturbations in drug efficacy and toxicity [2]. As such, much interest is focused on the identification, characterization and regulation of human CYPs and their contribution to metabolism within particular populations. Characterization of the involvement of the major hepatic CYPs (CYP1A2, 2C9, 2C19, 2D6 and 3A4) in drug metabolism was traditionally conducted using an array of methodologies including regression analysis with a panel of human liver microsomes (HLM), chemical and/or immuno-inhibition of the particular oxidative biotransformation and confirmation with recombinant CYPs, where available. Following the promise shown in early studies [3], advances in recombinant technology have seen a recent trend towards prioritizing the use of recombinant CYPs in such analyses [4, 5].
In general, such studies use human CYPs and HLM to provide a relative activity factor (RAF) for a specific prototypic/probe CYP reaction. Assays may be conducted under various conditions, for example with the velocity of the reaction (v) near Vmax, or using an intrinsic clearance, CLint, estimation [6–10]. These RAFs may then be used to ascertain the relative contribution of individual CYPs to the metabolism of a new molecular entity (NME). Alternatively, individual CYP CLint estimates may be scaled to provide a HLM equivalent based on the documented relative abundance of each hepatic isoform [5].
The aim of this study was to compare and contrast the utility of the relative abundance and activity approaches with the use of inhibitory monoclonal antibodies in the identification of human CYP enzymology. The experiments were also designed to reflect methodologies currently compatible with enhanced throughput drug discovery.
Methods and materials
Chemicals
All chemicals and reagents used were of the highest available commercial grade. Dextromethorphan, diclofenac, diltiazem (±)-metoprolol, phenacetin (±)-propranolol, testosterone and β-nicotinamide adenine dinucleotide phosphate, reduced form (β-NADPH) were purchased from Sigma Chemical Co. (Poole, Dorset, UK). (±)-Verapamil was purchased from Aldrich Chemical Co. Ltd. (Gillingham, UK). Bufuralol and S-mephenytoin were purchased from Ultrafine Chemicals (Manchester, UK). Omeprazole was synthesized at AstraZeneca R&D Charnwood (Loughborough, UK).
Source of cytochrome P450
Escherichia coli membranes expressing human CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 were purchased from CYPex (Dundee, UK). HLM (pooled with respect to CYP phenotype) were purchased from In Vitro Technologies (Baltimore, MD, USA) and have been described previously [5]. In Vitro Technologies obtain human tissue only from approved sources within the USA. Pooling by phenotype rather than genotype may confound interpretation of activities assigned to polymorphic enzymes such as CYP2C9/19 and CYP2D6.
CYP CLint determination
CYP CLint determination was performed using enhanced throughput methodology, similar to that reported previously [2, 5]. Initial substrate concentrations much lower than the respective Km of the reaction were used [5]. All compounds were incubated at 3 µm, except dextromethorphan (1 µm) and diclofenac (0.5 µm). (Based on previous kinetic analyses for dextromethorphan O-demethylation, the CYP2D6 CLint estimate would then be about 75% of the theoretical maximum [3].) The initial stock of all drug substrates was prepared in dimethyl sulfoxide at 100 times the incubation concentration. Thus, the final concentration of organic solvent in the incubation mixture was 1% v/v [5]. Escherichia coli membranes coexpressing individual CYPs and NADPH reductase (25–100 pmol of CYP ml−1 final concentration) [3, 5] were preincubated with substrate for 5 min at 37°C. Reactions were initiated by the addition of NADPH (1 mm final concentration) and 50-µl aliquots were taken at 0, 5, 10, 20 and 30 min and quenched with 100 µl ice-cold methanol. Samples were subsequently frozen for 1 h at −20°C, and then centrifuged at 2000 g for 20 min. The resultant supernatants were removed and transferred into HPLC vials prior to analysis.
Monoclonal antibody inhibition of CYP metabolism
MAbs used in this study have been shown previously to be specific for the following human CYP isoforms: CYP1A2 [11], CYP2C9 [12], CYP2C19 [12], CYP2D6 [13] and CYP3A4/5 [14]. MAbs (150 µg ascites fluid : mg microsomal protein) were preincubated with HLM (1 mg ml−1 final concentration) for 5 min at 37°C prior to the addition of substrates. Incubations were performed at identical concentrations to those in the CYP CLint determination. The mAb concentrations were chosen to be saturating for both HLM and rCYP reactions based on previous studies. The microsomal concentration selected was based on a compromise between assay sensitivity and nonspecific binding considerations. Because of the physicochemical properties of the compounds studied, some effects of the latter on apparent CLint may be anticipated, although this was not addressed directly in this study. Reactions were initiated with the addition of NADPH (1 mm final concentration), aliquots were taken at 0, 5, 10, 20 and 30 min and samples were extracted as in CYP CLint assays. Substrates were also incubated with HLM alone to determine the CLint without mAbs. Control HLM CLint values were of a similar magnitude to those reported previously [3, 5]. Percentage inhibition of metabolism was calculated as: 100 × (1 − CLint in the presence of the specific mAb)/CLint obtained with control mAb.
Analysis of recombinant CYP and human liver microsome samples
The majority of sample analysis was performed using a Micromass ZMD single quadrupole mass spectrometer using an HP1100 high performance liquid chromatography (HPLC) system for separation. Electrospray ionization was used in all mass spectrometry methods. Positive ion mode was used in parent loss analysis of bufuralol (m/z 262.2), diltiazem (m/z 415.2), dextromethorphan (m/z 272.2), metoprolol (m/z 268.2), omeprazole (m/z 346.1), phenacetin (m/z 180.1), propranolol (m/z 260.2), testosterone (m/z 289.2) and verapamil (m/z 455.3).
Chromatographic separation was obtained using a Symmetry Shield™ RP8 (4.6 × 50 mm, 3.5 µm) column (Waters, Watford, UK) using 20 µl of each extracted sample. The mobile phase consisted of water with 0.1% (v/v) formic acid with the organic phase being methanol containing 0.1% (v/v) formic acid. All chromatography was performed using a generic gradient (t = 0 min % organic = 10, t = 0.5 min % organic = 10, t = 4 min % organic = 100, t = 5 min % organic = 100, t = 5.1 min % organic = 10, total runtime = 5.5 min). The flow rate was set at 1.5 ml min−1, and mobile phase was introduced into the source at 0.4 ml min−1.
Analysis of 4′-hydroxymephenytoin was conducted on a Micromass Quatro Ultima triple quadrapole using an Alliance HT Waters 2790 HPLC system for separation. Analysis was by multiple reaction monitoring (MRM) using positive-ion mode. 4′-Hydroxymephenytoin was detected monitoring the transition 235.3 > 150 using a cone voltage of 30 V and a collision energy of 15 eV. Chromatographic separation was achieved using a Symmetry® C8 (2.9 × 20 mm, 5 µm) column (Waters) with the following gradient: t = 0 min % organic = 10, t = 0.1 min % organic = 10, t = 0.5 min % organic = 100, t = 1 min % organic = 100, t = 1.2 min % organic = 10, t = 1.7 min % organic = 10, total runtime = 2 min. The organic and mobile phases were the same as in the analysis of previous compounds. The flow rate was set at 1.5 ml min−1, and the mobile phase was introduced into the source at 0.4 ml min−1. The limit of quantification of the assays was 2 ng ml−1.
Analysis of diclofenac samples was conducted by HPLC using an HP1100 system. A Symmetry Shield™ RP8 (4.6 × 50 mm, 3.5 µm) cartridge (Waters) and a mobile phase of 0.025% (w/v) ammonium acetate and acetonitrile was used with the following gradient: t = 0 min % organic = 20, t = 7 min % organic = 60, t = 7.1 min % organic = 20, t = 8 min, total runtime = 8 min. The flow rate was 1.5 ml min−1 and u.v. detection of diclofenac was performed at 275 nm. For all analyses quoted, the intra- and inter-day coefficients of variation were < 10%.
Data analysis
Throughout this study, several approaches have been adopted to calculate intrinsic clearance (CLint). For the majority of compounds, CLint was determined by quantification of the specific rate of loss of parent compound [5]. The low turnover of more stable compounds (e.g. tolbutamide, S-mephenytoin) necessitated that the CLint was determined from the appearance of the metabolite [5]. The CLint for ethoxyresorufin O-de-ethlyation was determined from the expression Vmax/Km as reported previously [3]. The contribution of individual CYP to HLM CLint (abundance method) was estimated as detailed previously [5] using the immunoquantified concentrations of CYP isoforms [15] shown in Table 1 and a value of 320 pmol mg−1 as an average total CYP content in HLM [16].
Table 1.
Relative activity factor and immunoquantified estimates for major human CYPs in human liver microsomes.
| Percentage total CYP | |||||||
|---|---|---|---|---|---|---|---|
| Compound | Isoform | HLM CLintµl min−1mg−1 | CYP CLintµl min−1pmol−1 | AZ RAF pmol mg−1 | RAF | Immuno | References |
| Ethoxyresorufin | CYP1A2 | 40 ± 3 | 0.6 ± 0.2 | 67 | 21 | 13 | [15] |
| Tolbutamide | CYP2C9 | 0.7 ± 0.2 | 0.014 ± 0.002 | 50 | 16 | 20 | [15] |
| S-mephenytoin | CYP2C19 | 5 ± 1 | 0.66 ± 0.12 | 8 | 2 | 4 | [17, 29] |
| Dextromethorphan | CYP2D6 | 22 ± 1 | 1.97 ± 0.20 | 11 | 3 | 2 | [15] |
| Testosterone | CYP3A4 | 35 ± 2 | 0.42 ± 0.10 | 83 | 26 | 30 | [15] |
Data are mean ± s.d. of three individual experiments. AZ RAF, AstraZeneca-derived relative activity factor; immuno, immunoquantified (to which references provided relate).
The contribution of individual CYP to HLM CLint using the RAF method was estimated by the method recommended by Nakajima et al.[10], as adopted previously [5]. The relative merits of this approach have been discussed [10] and highlight potential differences in Km or Vmax between CYP expression systems and HLM. The authors are aware that this study only demonstrated the suitability of this method for CYP1A2, CYP2D6 and CYP3A4 reactions. However, CLint values determined in both HLM and recombinant CYP were used to calculate the contribution of individual CYP to HLM CLint using the following equation: Contribution of CYP (%) = (individual CYP Clint × RAF)/(ΣCYP Clint × RAF) × 100. RAFs for CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 were determined using the probe drugs shown in Table 1.
Results
Five compounds were selected from the literature to represent specific probe substrates for five major CYP isoforms, namely ethoxyresorufin (CYP1A2), tolbutamide (CYP2C9), S-mephenytoin (CYP2C19), dextromethorphan (CYP2D6) and testosterone (CYP3A4). CLint values were determined for each of the probe substrates using both pooled HLM (prepared from 15 individual livers) and the appropriate recombinant CYP cell line (Table 1). HLM RAFs were then calculated for each probe reaction and ranged from 8 pmol mg−1 for S-mephenytoin metabolism to 83 pmol mg−1 for testosterone metabolism.
The % contributions of CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 metabolism in human hepatic microsomes were calculated using the RAF approach (assuming 320 pmol of CYP mg−1 of microsomal protein) and compared with literature values determined using immunoquantified concentrations of CYP isoforms (Table 1). Both RAF and immunological methods (Fig. 1) gave comparable values for CYP1A2, CYP2C9, CYP2D6 and CYP3A4/5. However, the RAF estimates for the content of CYP2C19 in HLM were two-fold lower than the previous determination using immunoquantification techniques [17].
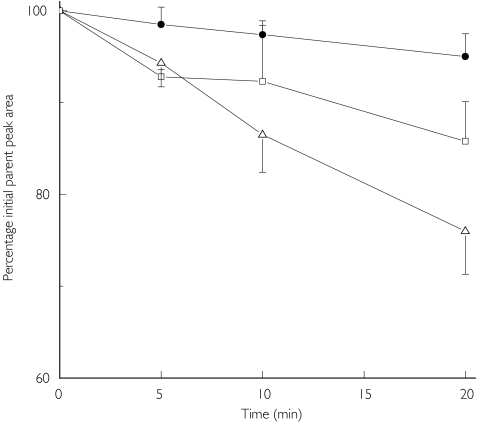
Figure 1.
The characterization of CYP-mediated propranolol metabolism using HLM and mAb analysis. Aliquots were taken at 0, 5, 10 and 20 min, and the amount of propranolol remaining in the incubation media expressed as the peak area at each time normalized to that observed at t = 0 min. The data (mean ± s.d.) represent propranolol clearance by HLM in the presence of CYP1A2 mAb (□), CYP2D6 mAb (•) and control mAb (▵) in three individual experiments.
Tables 2 and 3 show the results obtained when CYP CLint values determined for 14 drugs in the authors’ laboratory were analysed using abundance, RAF and mAb methods. Table 2 reflects the potential for a given drug to be metabolized by the individual CYPs studied, whereas Table 3 shows their actual contribution in the HLM pool studied. The reaction phenotyping by each of the three methods used in this study provided a similar definition of the individual major CYPs for each substrate. A major discrepancy was observed for phenacetin O-de-ethylation. This reaction has been shown to be metabolized by most hepatic CYPs but is a selective CYP1A2 probe at low substrate concentrations. The contribution of CYP2C19, thought to be the major lower affinity phenacetin O-de-ethylase, appeared to be overestimated based on data using recombinant CYPs, as suggested from data in Table 1.
Table 2.
Mean % contribution of individual CYPs to oxidative metabolism determined using average % content (APC) and relative activity factor (RAF) methods
| Percentage contribution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CYP1A2 | CYP2C9 | CYP2C19 | CYP2D6 | CYP3A4 | ||||||
| Compound | APC | RAF | APC | RAF | APC | RAF | APC | RAF | APC | RAF |
| Phenacetin | 44 | 58 | 5 | 4 | 22 | 14 | 2 | 3 | 27 | 21 |
| Naproxen* | 49 | 28 | 50 | 71 | 1 | 2 | – | – | – | – |
| Diclofenac | – | – | 100 | 100 | – | – | – | – | – | – |
| Omeprazole | – | – | – | – | 68 | 60 | – | – | 32 | 40 |
| Bufuralol | – | – | – | – | – | – | 100 | 100 | – | – |
| Metoprolol | – | – | – | – | – | – | 100 | 100 | – | – |
| Propranolol | 26 | 27 | – | – | 15 | 6 | 59 | 67 | – | – |
| Diltazem | – | – | – | – | 6 | 5 | – | – | 94 | 95 |
| Verapamil | – | – | – | – | – | – | – | – | 100 | 100 |
| Tolbutamide | – | – | 70 | 72 | 30 | 28 | – | – | – | – |
| Dextromethorphan | – | – | – | – | 14 | 10 | 86 | 90 | – | – |
| Testosterone | – | – | – | – | – | – | – | – | 100 | 100 |
| Diazepam* | – | – | – | – | 94 | 95 | – | – | 6 | 5 |
| Ibuprofen* | – | – | 90 | 91 | 10 | 9 | – | – | – | – |
Original raw data taken from [3] or [5]. APC and RAF values represent mean derived from three individual experiments. Naproxen provided a RAF for tolbutamide CYP2C9 (45 pmol mg−1), metoprolol for dextromethorphan CYP2D6 (6 pmol mg−1) and verapamil for testosterone CYP3A4 (98 pmol mg−1). –, Not detectable.
Table 3.
Percentage inhibition of oxidative metabolism by HLM CYPs using mAbs.
| Percentage inhibition | ||||||
|---|---|---|---|---|---|---|
| Compound | Control HLM Clint(µl min−1mg−1) | CYP1A2 | CYP2C9 | CYP2C19 | CYP2D6 | CYP3A4/5 |
| Phenacetin | 9 ± 2 | 83 ± 4 | – | – | – | – |
| Diclofenac | 80 ± 18 | – | 88 ± 1 | – | – | – |
| Omeprazole | 12 ± 1 | – | – | 39 ± 4 | – | 74 ± 11 |
| Bufuralol | 20 ± 4 | – | – | – | 100, 94 | – |
| Metoprolol | 4 ± 2 | – | – | – | 100 ± 0 | – |
| Propranolol | 11 ± 2 | 35 ± 9 | – | – | 83 ± 4 | – |
| Diltiazem | 27 ± 5 | – | – | – | – | 75 ± 4 |
| Verapamil | 138 ± 35 | – | – | – | – | 67 ± 10 |
| Dextromethorphan | 22 ± 1 | – | – | – | 77, 91 | 18, 5 |
| Testosterone | 35 ± 2 | – | 13 ± 7 | – | – | 50 ± 24 |
Values represent mean ± s.d. of three experiments or data from individual experiments. –, No consistent effect with mAb observed.
The mAb approach was unable to detect relatively minor contributions to metabolism (< 15%) using this enhanced throughput methodology. Both abundance and RAF methods estimated the contribution of CYP2C19- and CYP3A4-mediated metabolism of omeprazole to be approximately 65% and 35%, respectively. In contrast, the mAb method determined the relative importance of these isoforms to be approximately 2 : 1 in favour of CYP3A4.
The quantitative contribution to drug metabolism catalysed by CYP1A2, CYP2C9, CYP2D6 and CYP3A4 was also similar (generally, within two-fold) for abundance and RAF methods (Table 2). However, the relative importance of metabolism by CYP2C19 was overestimated by the abundance method compared with both RAF and mAb approaches. In the majority of cases, the degree of inhibition observed using mAbs reflected accurately the relative contribution of each CYP obtained by both abundance and RAF techniques. However, although drugs such as testosterone and verapamil were correctly identified as CYP3A4 substrates using mAbs, the degree of inhibition was only 50–75% of the contribution determined using other methods. The use of higher concentrations of antibody (300 µg ascites fluid per mg microsomal protein) in inhibition experiments resulted in a degree of inhibition comparable to estimates of the % contribution of the particular CYP (for example,> 83% for verapamil metabolism, data not shown).
Discussion
The large number of drug classes that interact with human CYPs necessitates a thorough understanding of this enzyme superfamily to expedite rapid drug development by pharmaceutical companies. The recent advances in recombinant DNA technology have allowed for significant improvements in evaluating the in vitro role of human CYPs in drug metabolism [18]. In particular, the relationship between CYP-mediated metabolism by recombinant CYPs and HLM has received considerable attention [4, 5, 18].
To examine the potential use of abundance, RAF and mAb methods in early drug discovery programmes, reaction phenotyping of a range of drug substrates was performed with enhanced throughput technology using each of the three methods. Throughout the majority of this investigation, the CLint of drugs was determined by monitoring loss of parent drug, as advocated previously [19, 20]. Although this method is less sensitive and selective than the determination of CLint values for specific metabolite formation, it was employed to mimic techniques commonly used in early drug discovery, where tight deadlines often prohibit extensive method development and synthesis of authentic metabolite standards.
The probe substrates (ethoxyresorufin, tolbutamide, S-mephenytoin, dextromethorphan and testosterone) used to create RAFs for CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4, respectively, have been used widely [3, 5, 7, 8, 21]. Traditionally many investigators have estimated RAFs using probe substrates under Vmax conditions in both recombinant CYPs and HLM [6–9]. However, CLint estimates have been used to determine RAFs in present study based on the work of Nakajima et al.[10], who suggested that this method more accurately mimicked the situation in vivo. RAFs determined for CYP2C9 and CYP2C19 were within previous literature ranges determined using the same probe substrates [7, 8, 22]. However, the RAFs for CYP1A2, CYP2D6 and CYP3A4 were several times lower than those obtained by previous investigators [22, 23]. The relative activity approach may depend on the index reaction used to create RAFs for each CYP isoform and the expression system employed [10]. To investigate this phenomenon further, several compounds were used to determine interprobe variation for each major CYP isoform. With the exception of CYP2D6 (where interprobe variation was four-fold) all other isoform selective probe reactions gave RAFs within approximately two-fold of each other, which agrees favourably with the literature [24].
Although the use of relative abundance and activity methods for reaction phenotyping CYPs has been compared for a limited number of individual drugs previously [22, 24, 25], a systematic evaluation of the two techniques for a substantial number of drugs has yet to be performed. The considerable potential use of mAbs in drug discovery studies has become apparent recently [26, 27]. The quantitative contribution to drug metabolism catalysed by CYP1A2, CYP2C9, CYP2D6 and CYP3A4 was similar using average % content (APC) and RAF methods. This finding supports that of Yu and Haining [25], who demonstrated that the CYP2C9, CYP2D6 and CYP3A4-mediated metabolism of dextromethorphan was comparable using each method. Several investigators [22, 24] have reported the predicted contribution of CYP1A2 to drug biotransformation using the RAF approach to be 5–20-fold higher than APC methods. Venkatakrishnan et al.[24] suggested that such differences may be attributed to differing expression levels of NADPH : CYP oxidoreductase in recombinant CYPs and HLM. Yamazaki et al.[26] have demonstrated that the coexpression of recombinant CYPs (in particular CYP3A4) with cytochrome b5 may also aid in comparisons between some recombinant systems and HLM. Therefore, it is tempting to speculate that the func-tional coupling of NADPH : CYP oxidoreductase : cytochrome b5 in the HLM and recombinant CYPs used in this study were equivalent. Interestingly, Venkatakrishnan et al.[24] also observed a discrepancy in accessory protein content between HLM and recombinant CYP2C19. However, differences in predicted CYP2C19-mediated metabolism using APC and RAF approaches were minimal. Another explanation could be that the immunoquantified concentration of CYP2C19 (commonly quoted as 4%) might be an overprediction, and that a value of 1–2% may be more appropriate (Table 1[27]). The presence of CYP2C19 poor metabolizing livers in the HLM used here may also confound interpretation of the data.
In addition to the APC and RAF approaches, our data clearly demonstrate the potential use of mAbs to quantitatively reaction phenotype HLM for CYP-mediated processes. The major differences using mAb and APC/RAF techniques in the predictive contribution of CYP2C19 to phenacetin O-de-ethylase and CYP2C19 and CYP3A4 to omeprazole metabolism may be characteristic of the HLM batch used [28]. Therefore, further work investigating individual HLM samples may be required. Relatively minor CYP contributions to drug CLint (where % contribution was < 15%) were not detected consistently with mAbs in this study, in which compounds were incubated at substrate concentration of less than the Km for the major, high-affinity pathway under the conditions chosen. However, previous investigations using much higher substrate concentrations have shown that mAbs have the potential to detect contributions from more minor CYPs [12, 13], although the physiological/pharmacological significance of this is debatable. The differing incubation conditions may also explain the lower % inhibition of certain CYP3A4 reactions studied by us compared with previous studies (often conducted under Vmax conditions) [24, 27].
This report has evaluated APC, RAF and mAb approaches towards the CYP reaction phenotyping using 14 drug substrates. The successful determination of the contribution of the major CYP-mediated biotransformations by the three approaches supports further evaluation of APC, RAF and mAb techniques in early drug discovery.
References
- 1.Bertz RJ, Granneman GR. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokinet. 1997;32:210–258. doi: 10.2165/00003088-199732030-00004. [DOI] [PubMed] [Google Scholar]
- 2.Riley RJ. The potential pharmacological and toxicological impact of P450 screening. Curr Opin Drug Discovery Dev. 2001;4:45–54. [PubMed] [Google Scholar]
- 3.McGinnity DF, Griffin SJ, Moody GC, et al. Rapid characterization of the major drug-metabolizing human hepatic cytochrome P-450 enzymes expressed in Escherichia coli. Drug Metab Dispos. 1999;27:1017–1023. [PubMed] [Google Scholar]
- 4.Eddershaw PJ, Dickins M. Advances in in vitro drug metabolism screening. Pharm Sci Technol Today. 1999;2:13–19. doi: 10.1016/s1461-5347(98)00108-4. [DOI] [PubMed] [Google Scholar]
- 5.McGinnity DF, Parker AJ, Soars M, Riley RJ. Automated definition of the enzymology of drug oxidation by the major human drug metabolizing cytochrome P450s. Drug Metab Dispos. 2000;28:1327–1334. [PubMed] [Google Scholar]
- 6.Crespi CL. Xenobiotic-metabolizing human cells as tools for pharmacological and toxicological research. Adv Drug Res. 1995;26:179–235. [Google Scholar]
- 7.Kobayashi K, Chiba K, Yagi T, et al. Identification of cytochrome P450 isoforms involved in citalopram N-demethylation by human liver microsomes. J Pharmacol Exp Ther. 1997;280:927–933. [PubMed] [Google Scholar]
- 8.Venkatakrishnan K, Von Moltke LL, Greenblatt DJ. Relative quantities of catalytically active CYP2C9 and 2C19 in human liver microsomes: application of the relative activity factor approach. J Pharm Sci. 1998;87:845–853. doi: 10.1021/js970435t. [DOI] [PubMed] [Google Scholar]
- 9.Von Moltke LL, Greenblatt DJ, Grassi JM, et al. Multiple human cytochromes contribute to biotransformation of dextromethorphan in vitro: role of CYP2C9, CYP2C19, CYP2D6, and CYP3A. J Pham Pharmacol. 1998;50:997–1004. doi: 10.1111/j.2042-7158.1998.tb06914.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakajima M, Nakamura S, Tokudome S, Shimada N, Yamazaki H, Yokoi T. Azelastine N-demethylation by cytochrome P-450 (CYP) 3A4, CYP2D6, and CYP1A2 in human liver microsomes: evaluation of approach to predict the contribution of multiple CYPs. Drug Metab Dispos. 1999;27:1381–1391. [PubMed] [Google Scholar]
- 11.Yang TJ, Sai T, Krausz KW, Gonzalez FJ, Gelboin HV. Inhibitory monoclonal antibodies to human cytochrome P450 1A2: analysis of phenacetin O-de-ethylation in human liver. Pharmacogenetics. 1997;8:375–382. doi: 10.1097/00008571-199810000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Krausz KW, Goldfarb I, Buters JTM, Yang TJ, Gonzalez FJ, Gelboin HV. Monoclonal antibodies specific and inhibitory to human cytochromes P450 2C8, 2C9, and 2C19. Drug Metab Dispos. 2001;29:1410–1423. [PubMed] [Google Scholar]
- 13.Gelboin HV, Krausz KW, Shou M, Gonzalez FJ, Yang TJ. A monoclonal antibody inhibitory to human P450 2D6: a paradigm for use in combinatorial determination of individual P450 role in specific drug tissue metabolism. Pharmacogenetics. 1997;7:469–477. doi: 10.1097/00008571-199712000-00005. [DOI] [PubMed] [Google Scholar]
- 14.Gelboin HV, Krausz KW, Goldfarb I, et al. Inhibitory and non-inhibitory monoclonal antibodies to human cytochrome P450 3A3/4. Biochem Pharmacol. 1995;50:1841–1850. doi: 10.1016/0006-2952(95)02077-2. [DOI] [PubMed] [Google Scholar]
- 15.Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals. Studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;170:414–423. [PubMed] [Google Scholar]
- 16.Iwatsubo T, Hirota N, Ooie T, et al. Prediction of in vivo drug metabolism in the human liver from in vitro metabolism data. Pharmacol Ther. 1997;73:5147–5171. doi: 10.1016/s0163-7258(96)00184-2. [DOI] [PubMed] [Google Scholar]
- 17.Wester MR, Lasker JM, Johnson EF, Raucy JL. CYP2C19 participates in tolbutamide hydroxylation by human liver microsomes. Drug Metab Dispos. 2000;28:354–359. [PubMed] [Google Scholar]
- 18.Rodrigues AD. Integrated cytochrome P450 reaction phenotyping: attempting to bridge the gap between cDNA-expressed cytochromes P450 and native human liver microsomes. Biochem Pharmacol. 1999;57:465–480. doi: 10.1016/s0006-2952(98)00268-8. [DOI] [PubMed] [Google Scholar]
- 19.Carlile DJ, Stevens AJ, Ashforth EIL, Waghela D, Houston JB. In vivo clearance of ethoxycoumarin and its prediction from in vitro systems. Use of drug depletion and metabolite formation methods in hepatic microsomes and isolated hepatocytes. Drug Metab Dispos. 1998;26:216–221. [PubMed] [Google Scholar]
- 20.Suzuki A, Iida I, Tanaka F, et al. Identification of human cytochrome P-450 isoforms involved in metabolism of R(+)- and S(-)-gallopamil: utility of in vitro disappearance rate. Drug Metab Dispos. 1999;27:1254–1259. [PubMed] [Google Scholar]
- 21.Waxman DJ, Attisano C, Guengerich FP, Lapenson DP. Human liver microsomal steroid metabolism: identification of the major microsomal steroid hormone 6 beta-hydroxylase cytochrome P-450 enzyme. Arch Biochem Biophys. 1988;263:424–436. doi: 10.1016/0003-9861(88)90655-8. [DOI] [PubMed] [Google Scholar]
- 22.Stormer E, Von Moltke LL, Greenblatt DJ. Scaling drug biotransformation data from cDNA-expressed cytochrome P-450 to human liver: a comparison of relative activity factors and human liver abundance in studies of mirtazapine metabolism. J Pharmacol Exp Ther. 2000;295:793–801. [PubMed] [Google Scholar]
- 23.Venkatakrishnan K, Von Moltke LL, Greenblatt DJ. Application of the relative activity factor approach in scaling from heterologously expressed cytochromes P450 to human liver microsomes: studies on amitriptyline as a model substrate. J Pharmacol Exp Ther. 2001;297:326–337. [PubMed] [Google Scholar]
- 24.Venkatakrishnan K, Von Moltke LL, Court MH, Harmatz JS, Crespi CL, Greenblatt DJ. Comparison between cytochrome P450 (CYP) content and relative activity approaches to scaling from cDNA-expressed CYPs to human liver microsomes: ratios of accessory proteins as sources of discrepancies between the approaches. Drug Metab Dispos. 2000;28:1493–1504. [PubMed] [Google Scholar]
- 25.Yu A, Haining R. Comparative contribution to dextromethorphan metabolism by cytochrome P450 isoforms in vitro. Can dextromethorphan be used as a dual probe for both CYP2D6 and CYP3A activities? Drug Metab Dispos. 2001;29:1514–1520. [PubMed] [Google Scholar]
- 26.Yamazaki H, Nakajima M, Nakamura M, et al. Enhancement of cytochrome P-450 3A4 catalytic activities by cytochrome b5 in bacterial membranes. Drug Metab Dispos. 1999;27:999–1004. [PubMed] [Google Scholar]
- 27.Shou M, Lu T, Krausz W, et al. Use of inhibitory monoclonal antibodies to assess the contribution of cytochromes P450 to human drug metabolism. Eur J Pharmacol. 2000;394:199–209. doi: 10.1016/s0014-2999(00)00079-0. [DOI] [PubMed] [Google Scholar]
- 28.Yamazaki H, Inoue K, Shaw PM, Checovich WJ, Guengerich FP, Shimada T. Different contributions of cytochrome P450 2C19 and 3A4 in the oxidation of omeprazole by human liver microsomes: effects of contents of these two forms in individual human samples. J Pharmacol Exp Ther. 1997;283:434–442. [PubMed] [Google Scholar]
- 29.Lasker JM, Wester MR, Aramsombatdee E, Raucy JL. Characterization of CYP2C19 and CYP2C9 from human liver: respective roles in microsomal tolbutamide, S-mephenytoin, and omeprazole hydroxylations. Arch Biochem Biophys. 1998;353:16–28. doi: 10.1006/abbi.1998.0615. [DOI] [PubMed] [Google Scholar]