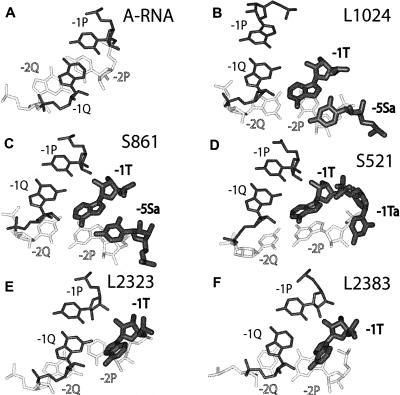
FIGURE 3.
The displacement of base pair [− 2P;− 2Q] with respect to [− 1P;− 1Q] in the G-ribo-based pseudoknots. All structures are aligned by the position of nucleotide − 1Q. Nucleotides − 2P and − 2Q are white, while all other nucleotides are black. Nucleotides − 1T, − 5Sa (only in L1023 and S861), and − 1Ta (only in S521), which are involved in the stabilization of the position of nucleotide − 2P, are shown thick. Compared to the juxtaposition of two consecutive base pairs in a regular RNA double helix (A), base pair [− 2P;− 2Q] in the G-ribo wrenches (B–D) is rotated for about 50° around atom O3′ of − 2Q in the direction of the minor groove and is additionally shifted for about 4 Å. In motifs L2323 and L2383 forming the G-ribo ring (E,F), the displacement of base pair [− 2P;− 2Q] is smaller than in the G-ribo wrenches, and in L2323 (E), it is smaller than in L2383 (F). Due to this displacement, − 2P in all pseudoknots has lost its interaction with − 1P and becomes stacked to − 1T. Additional stabilization of the − 2P position in the G-ribo wrenches is provided through the interaction of its ribose with either − 5Sa or − 1Ta. Uridine − 5Sa, when it exists, forms a Hoogsteen base pair with adenosine − 1T.