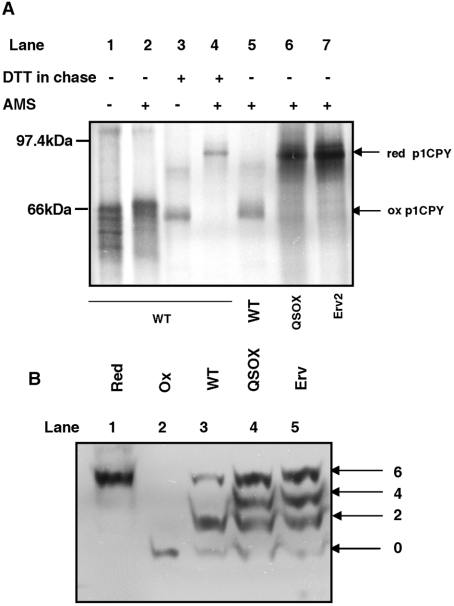
Figure 6. Redox states of CPY and PDI.
(A) BY4742 wild-type (WT) cells were radiolabelled for 10 min in the absence (lanes 1 and 2) or presence of 10 mM DTT (lanes 3 and 4) to detect the fully oxidized and fully reduced p1CPY respectively. BY4742 (WT), ero1Δ PGAL1-hQSOX1a (QSOX) and ero1Δ PGAL1-ERV2 (Erv) were radiolabelled for 10 min in the presence of 5 mM DTT followed by a 30 min chase in the absence of DTT (lanes 5, 6 and 7). Cells were isolated at the end of the labelling and chase periods and proteins from cell pellets were TCA precipitated and solubilized in the presence (lanes 2 and 4–7) or absence (lanes 1 and 3) of 25 mM AMS prior to immunoisolation of CPY. Samples were resolved by non-reducing SDS/PAGE. The reduced (red) and oxidized (ox) forms of p1CPY are indicated. (B) TCA (20%) was added to logarithmically growing cultures to quench thiol/disulfide exchange and precipitate proteins. Free sulfhydryl groups were modified by resuspension in 5 mM Mal-PEG under denaturing conditions [3% SDS and 0.2 M Tris/HCl, (pH 8)]. Proteins were separated by SDS/PAGE and visualized by Western blotting using an anti-yeast PDI antibody. Apparent molecular mass shifts above the unmodified protein indicate the number of free sulfhydryl groups modified. The redox states of PDI in the parental BY4742 (WT), ero1Δ PGAL1-hQSOX1a (QSOX) and ero1Δ PGAL1-ERV2 (Erv) are compared with reduced (Red) and oxidized (Ox) controls that were treated with 10 mM DTT or 1 mM diamide before Mal-PEG modification.