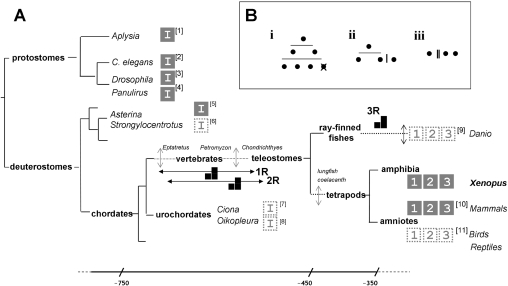
Figure 4. Molecular evolution of the Itpr gene family.
(A) A simplified triploblast phylogeny, drawn on the basis of the Protostomia (Lophotrochozoa–Ecdysozoa)–Deuterostomia classification to highlight the evolutionary diversification of the Itpr gene family. For clarity, certain nodes have been omitted and are represented by a broken line with a vertical arrow to indicate intermediate divergence of lineages. Approximate timings (millions of years) are shown on a non-linear scale (bottom). Proposed timings of WGD events (1R, 2R and 3R) are indicated on the appropriate branch of the phylogeny. Key model organisms (italicized), including those in which a single Itpr gene (‘I’) or multiple Itpr genes (subtype ‘1’, ‘2’ and ‘3’) have been confirmed by sequencing of a full-length clone from biological material (solid box), or predicted in part by genome annotation (dashed box) are shown. Accession numbers (square brackets) are as follows: ‘1’, DQ397517 (Aplysia californica); ‘2’, AJ243181 (Caenorhabditis elegans); ‘3’, D90403 (Drosophila melanogaster); ‘4’, AF055079 (Panilurus argus); ‘5’, AB071372 (Asterina pectinifera) [38]; ‘6’, XR_025864 (Strongylocentrotus purpuratus) ‘7’, ENSCING00000007322 (Ciona intestinalis) and ENSCSAVG00000006185 (Ciona savignyi); ‘8’, AAT47836 (Oikopleura dioica); ‘9’, XM_685965 and XP_693046 (Danio rerio); ‘11’, XM_414438, XM_001235612 and XM_418035 (Gallus gallus) [36]. For mammalian IP3R diversity (‘10’), see [14,16]. (B) Alternative gene duplication pedigrees leading to three Itpr paralogues are represented by (i) two WGD (2R, horizontal lines), followed by gene loss, (ii) one round of WGD (1R) followed by a more localized duplication (vertical line), or (iii) two successive local duplications.