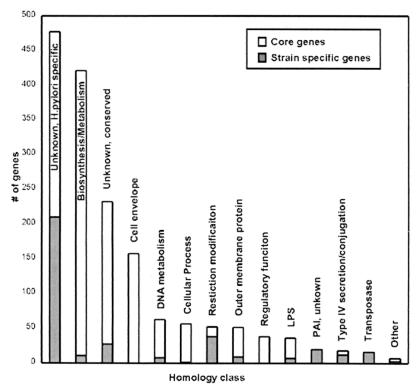
Figure 1.
Homology classes of H. pylori core and strain-specific genes. Each bar represents the number of strain-specific (gray) and core genes (white) in each of the following homology classes (number of genes/percentage of genes): unknown, H. pylori specific strain-specific (210/56%) core (267/21%); metabolism and biosynthesis, excluding DNA metabolism, strain-specific (11/3%) core (410/32%) strain specific; unknown, conserved strain-specific (27/7%) core (205/16%); cell envelope, excluding OMP and lipopolysaccharide (LPS), strain-specific (0/0%) core (157/12%); DNA metabolism, strain-specific (8/2%) core (54/4%); cellular process, strain-specific (1/0.3%) core (55/4%); restriction modification, strain-specific (38/10%) core (14/1%); OMP, strain-specific (9/2%) core (42/3%); regulatory function, strain-specific (0/0%) core (38/3%); LPS biosynthesis, strain-specific (7/2%) core (29/2%); PAI, unknown function, strain-specific (20/6%) core (0/0%); type IV secretion and conjugation, strain-specific (12/3%) core (6/0.5%); transposase, strain-specific (16/4%) core (0/0%); other, strain-specific (3/0.8%) core (4/0.3%).