FIG. 1.
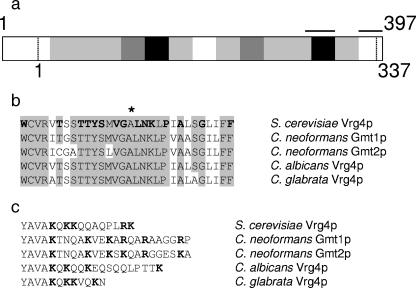
GDP-Man transporters from fungi. (a) The level of identity between the predicted protein sequence of C. neoformans Gmt1 and S. cerevisiae Vrg4 is indicated by the shading of each 25-amino-acid segment: 0 to 25%, white; 26 to 50%, light gray; 51 to 75%, dark gray; 76 to 100%, black. Horizontal bars indicate the regions of sequence that are expanded in b (left bar) and c (right bar). Vertical dashed lines indicate the termini of the shorter Vrg4. (b) Shown are aligned GALNK regions of GDP-Man transporters encoded by genes from S. cerevisiae (VRG4) (GenBank accession number NP_011290), C. neoformans (GMT1 and GMT2), C. albicans (GenBank accession number AAK74075), and C. glabrata (GenBank accession number AAK51897). The alignment begins with the tryptophan at position 271 of Vrg4. The originally identified components of the sequence motif (17) are indicated in boldface type in the S. cerevisiae sequence, and * indicates the amino acid mutated to aspartic acid in the S. cerevisiae vrg4-2 mutant. (c) Shown are the C-terminal hydrophilic residues that follow a conserved hydrophobic stretch (partially shown, YAVA) of each transporter. Basic residues are indicated in boldface type.