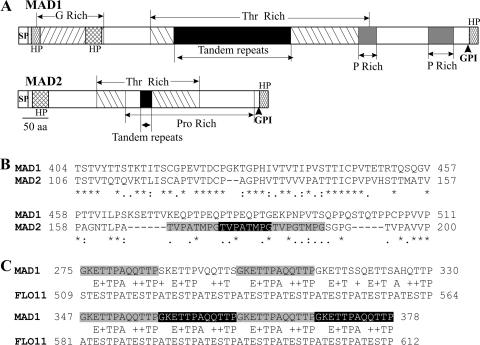
FIG. 1.
Structural features of MAD1 and MAD2. (A) Schematic structure of MAD1 and MAD2 showing glycine-, threonine-, and proline-rich regions. SP, signal peptide; GPI, glycosylphosphatidylinositol anchor site; HP, hydrophobic region. (B) ClustalX alignment of the conserved region in MAD1 and MAD2 (* indicates consensus sites). The MAD2 tandem repeats are shadowed. (C) Conservation between tandem repeat regions (shadowed) in MAD1 and C. albicans FLO11.