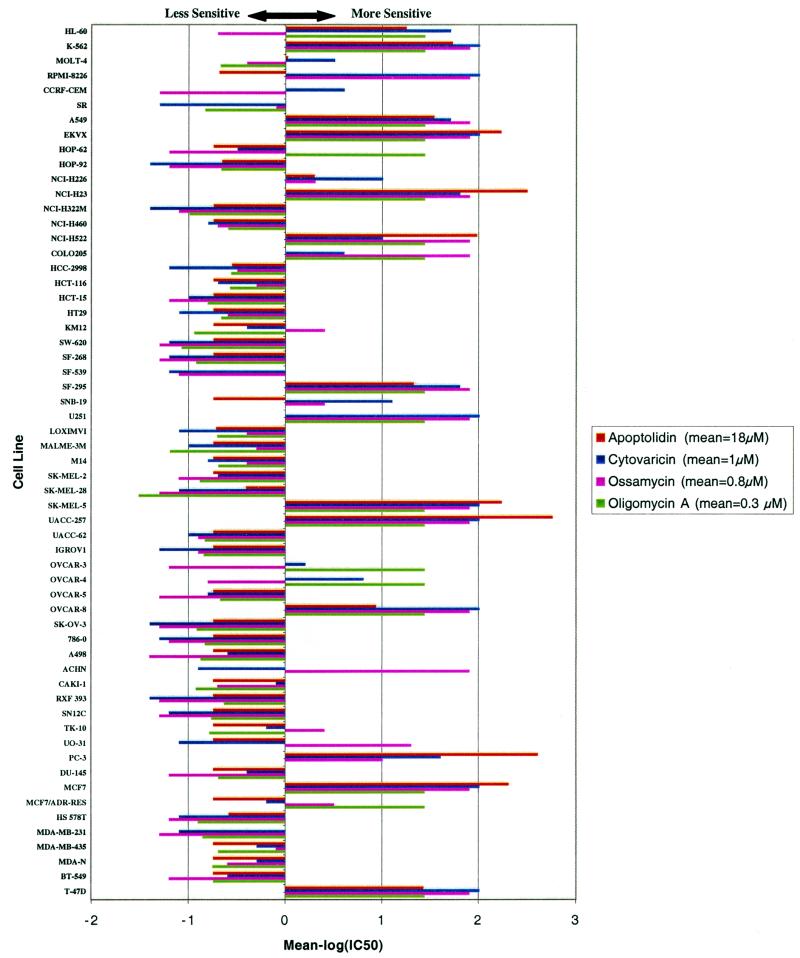
Figure 2.
Sensitivity profile for F0F1-ATPase inhibitors shown in Fig. 1 against the 60 human cancer cell lines of the National Cancer Institute (NCI-60). A powerful method to test the selectivity of an anticancer drug to various types of cancer is the sensitivity pattern among the NCI-60 (5, 6). By comparison of the pattern of sensitivity of a molecule to the approximately 70,000 molecules previously examined, one can often discover molecules of known mechanism of action that show high correlation to the compound, suggesting a similar mechanism of action (7). The mean IC50 values for apoptolidin, cytovaricin, oligomycin A, and ossamycin were 18 μM, 1 μM, 0.27 μM, and 0.8 μM, respectively. HL-50, K-562, MOLT-4, RPMI-8226, CCRF-CEM, and SR are tumor cell lines derived from human leukemias. A549, EKVX, HOP-62, HOP-92, NCI-H226, NCI-H23, NCI-H322 M, NCI-H460, and NCI-H522 are derived from lung cancers. COLO205, HCC-2998, HCT-116, HCT-15, HT29, KM12, and SW-620 are derived from colon cancers. SF-268, SF-539, SF-295, SNB-19, and U251 are derived from central nervous system cancers. LOXIMVI, MALME-3 M, M14, SK-MEL-2, SK-MEL-28, SK-MEL-5, UACC-257, and UACC-62 are derived from melanomas. IGROV1, OVCAR-3, OVCAR-4, OVCAR-5, OVCAR-8, and SK-OV-3 are derived from ovarian cancers. 786-0, A498, ACHN, CAKI-1, RXF 393, SN12C, TK-10, UO-31, and PC-3 are derived from renal cancers. DU-145 is derived from a prostate cancer. MCF7, HS 578T, MDA-MB-231, MDA-MB-435, MDA-N, BT-549, and T-47D are derived from breast cancers. MCF7/ADR-RES is a multidrug-resistant derivative of MCF7.