Abstract
Drosophila trachealess (Trl), master regulator of tracheogenesis, has no known functional mammalian homolog. We hypothesized that genes similar to trachealess regulate lung development. Quantitative (Q)RT-PCR and immunostaining were used to determine spatial and temporal patterns of npas1 gene expression in developing murine lung. Immunostaining for α-smooth muscle actin demonstrated myofibroblasts, and protein gene product (PGP)9.5 identified neuroendocrine cells. Branching morphogenesis of embryonic lung buds was analyzed in the presence of antisense or sense oligodeoxynucleotides (ODN). Microarray analyses were performed to screen for changes in gene expression in antisense-treated lungs. QRT-PCR was used to validate the altered expression of key genes identified on the microarrays. We demonstrate that npas1 is expressed in murine embryonic lung. npas1 mRNA peaks early at Embryonic Day (E)10.5–E11.5, then drops to low levels. Sequencing verifies the identity of npas1 transcripts in embryonic lung. NPAS1 immunostaining occurs in nuclei of parabronchial mesenchymal cells, especially at the tracheal bifurcation. Arnt, the murine homolog of Tango (the heterodimerization partner for Trl) is also expressed in developing lung but at constant levels. npas1- or arnt-antisense ODN inhibit lung branching morphogenesis, with altered myofibroblast development and increased pulmonary neuroendocrine cells. On microarrays, we identify > 50 known genes down-regulated by npas1-antisense, including multiple genes regulating cell migration and cell differentiation. QRT-PCR confirms significantly decreased expression of the neurogenic genes RBP-Jk and Tle, and three genes involved in muscle development: β−ig-h3, claudin-11, and myocardin. Npas1 can regulate myofibroblast distribution, branching morphogenesis, and neuroendocrine cell differentiation in murine embryonic lung.
Keywords: branching morphogenesis, myofibroblasts, smooth muscle actin, cell migration, neuroendocrine cells
CLINICAL RELEVANCE
This research identifies NPAS1 as a novel transcription factor regulating branching morphogenesis in murine lung buds. NPAS1 antisense oligos inhibit branching via a mechanism involving arrested myofibroblast migration and neuroendocrine cell hyperplasia.
Lung morphogenesis is the result of complex interactions among multiple transcription factors, growth factors, and signaling pathways (1–4). Tracheal development in Drosophila melanogaster provides a simple model for investigating analogous genes involved in respiratory system development (5). Trachealess (trl) is the master regulatory gene because null mutation of trl leads to complete absence of the fly respiratory system (6). Trl also regulates downstream branching morphogenesis of the developing tracheal system (6, 7). Trl is a transcription factor (TF) with a basic helix-loop-helix (bHLH) domain and two “PAS” domains (6). Several other PAS domain genes have been cloned, including periodicity (per), single-minded (sim), and aryl hydrocarbon nuclear translocator (arnt), all of which collectively led to the designation of this family of TFs as “PAS” (per-arnt-sim) domain proteins. We became particularly interested in npas1 because this gene is closely homologous to Drosophila trl. Amino acid alignment scores to the Drosophila Trl amino acid sequence on a pam250 matrix are for 393 for mouse Npas1, versus 335 for mouse Sim2 versus 210 for Arnt. We are interested in Arnt because Tango, its fly homolog, is required for Trl function via formation of heterodimers (8, 9), leading to regulation of the migration of the first tracheoblasts. This signaling initiates a developmental cascade critical for Drosophila tracheogenesis (9). Breathless (Drosophila fibroblast growth factor [FGF] receptor [FGFR]), branchless (Drosophila FGF), and sprouty, an FGF signaling antagonist, function downstream from Trl and also regulate tracheal branching (9). Notch, delta, and enhancer of split mutants have pleiotropic effects, including tracheal defects and increased neurogenesis (9). Similarly, pleiotropic signaling mechanisms with a role in both lung and tracheal development include FGF10 (2), the Nkx2.1 (1), and Notch-1 (10).
Functional information is now available for several PAS proteins. Murine NPAS1 (11, 12) is a PAS protein expressed in the central nervous system (CNS), where it regulates complex behaviors (13–15). In previous studies in other laboratories, npas1 mRNA was not detected in either adult murine lung or whole embryos before Embryonic Day (E)13 using Northern blot analyses (12). Npas1-null mice did not have any apparent pulmonary phenotype, suggesting that either it has no role in lung development, or that the phenotype may occur only in certain genetic backgrounds, or that there is functional redundancy with other genes. However, there has been no known direct analysis of lung histopathology in npas1-null mice, nor any functional assay related to lung development in these mice. It is known that in vitro functional assays can be more sensitive than phenotypic analysis of knockout mice because short-term gene knock-down does not allow sufficient time for compensatory gene upregulation that can occur in knockout mice (16, 17). Similarly, in reports of npas2 and npas3-null mice, there has been no report of pulmonary abnormalities. Consistent with arnt being ubiquitously expressed, arnt-null mice are embryonic lethal before E14, but the hemizygous arnt-null mice do not have any obvious defects. Hypoxia-inducible factor (HIF)-1α and HIF-2α are additional PAS domain genes known to be of functional significance in lung vascular development (18, 19), but these are expressed in later fetal lung development (⩾ E15.5) when alveolar capillary development begins.
None of the potential TRL homologs has been previously demonstrated in vivo to be associated with a mammalian respiratory phenotype. In the present study, we use a sensitive in vitro functional assay to test the hypothesis that NPAS1 can regulate lung development.
This work was presented in part at the International Conferences of the American Thoracic Society in 2003 and 2004 (20, 21).
MATERIALS AND METHODS
RNA Analyses
Total RNA was isolated from developing lungs from timed-pregnant Swiss-Webster mice (Taconic Laboratories, Germantown, NY) using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA was synthesized with SuperScript-III First-strand Synthesis System (Invitrogen). For QRT-PCR, primer pairs were designed to span introns (see online supplement for primer sequences) using Primer Express-3.0 (Applied Biosystems, Foster City, CA). For real-time-PCR, cDNA from 200 ng RNA, 0.5 μM primer pair, and 0.5 μM probe (Integrated DNA Technologies, Coralville, IA) were combined in Taqman Universal PCR Master-Mix (Applied Biosystems) and run in ABI-PRISM 7,300 (Applied Biosystems) at 50°C for 2-min, 95°C for 10 min, then 45-cycles at 95°C for 15 s each and 60°C for 1 min. Sequences of primers and probes for QRT-PCR of npas1, sim2, arnt, and β-actin are given in Table E1 in the online supplement. Sequences of primers and probes for QRT-PCR of claudin 11, myocardin, β-ig-H, RBPsuh, Tle, npas1 and β-actin are given in Table E2.
Antisense Oligodeoxynucleotide Treatment of Developing Lungs
E11.5 embryonic lungs were cultured on Transwell inserts as described (10). Specific oligodeoxynucleotides (ODN) (primer sequences given in online supplement) were added to the media, changed daily. Cultures were incubated for 2–6 d. Lungs were photographed at 0 and 48 h.
Immunohistochemistry
Lungs were fixed in 4% paraformaldehyde and embedded in paraffin. Immunostaining for proliferating cell nuclear antigen (PCNA), smooth muscle actin (SMA), and protein gene product (PGP)9.5 was performed using the ABC method (Vector Laboratories, Burlingame, CA) as described (10). Anti-NPAS1 antibody (ProSci, Inc., Poway, CA) was used at 1:100. Anti-SMA murine monoclonal antibody (Dako Laboratories, Carpinteria, CA) was used at 1:125; anti-PGP9.5 rabbit polyclonal antibody (Ultraclone, Isle of Wight, UK) was used at 1:1,000. Slides were counterstained with methyl green.
RNA Microarray Analyses
Total RNA was prepared from five separate experiments in which E11.5 lung buds were treated with npas1-AS or -sense for 48 h. cDNAs were prepared and microarrays were hybridized and analyzed as detailed in the website for the Harvard Partners Center for Genomics and Genetics at http://www.hpcgg.org/News/MicroarrayDocument.pdf.
The bio-informatics pipeline for microarray data analysis and reporting was accomplished through integration with an analysis/workflow tool called GenePattern (http://www.broad.mit.edu/cancer/software/genepattern/). Normalized data for each array were exported to the Partners HPCGG Bioinformatics Server (http://portal.hpcgg.org), merged with updated gene annotation data for each probe set on the array, and downloaded for further data visualization and analysis.
RESULTS
QRT-PCR for NPAS1, Arnt, and Sim2
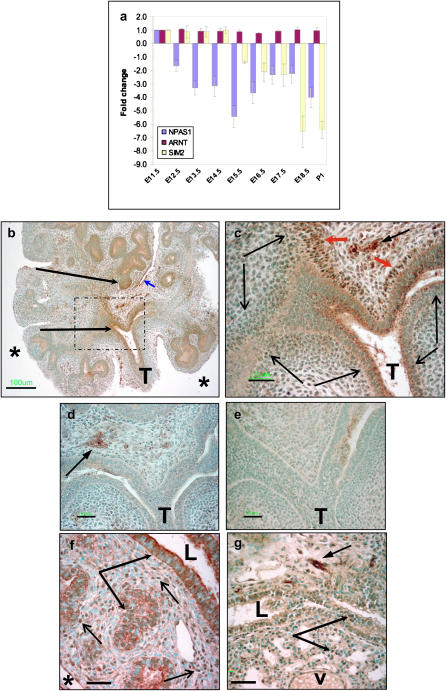
To determine temporal patterns and relative levels of gene expression, real-time quantitative RT-PCR (QRT-PCR) was performed for npas1 and arnt using mRNA from murine lung from E10–E18 and postnatal day 1 (P1) (Figure 1a). Transcript levels were normalized for β-actin and expressed relative to the unit level defined in E11.5 lung buds. Although arnt mRNA levels change minimally during development (Figure 1a), npas1 is expressed in embryonic lung during a specific developmental window. The level of npas1 mRNA is highest at E10.5–E11.5, then markedly declines at E12.5 (Figure 1a and data not shown). As a specificity control, we also performed QRT-PCR for sim2, because sim2-null mice have normal lung development (22). Sim2 is expressed at high levels from E10.5–E16.5, when mRNA levels drop markedly.
Figure 1.
Ontogeny of NPAS1 mRNA and protein expression in murine lung. (a) Real-time QRT-PCR was used to quantify npas1 mRNA levels in fetal lung, normalized to β-actin levels in the same RT reactions, and compared with levels at E11.5, which was defined as a unit level of one. For comparison, arnt and sim2 mRNA levels were also quantified. Whereas arnt mRNA levels are essentially unchanged from E11.5 to P1, npas1 drops at E12.5, and sim2 drops at E16.5. (b–e) Immunoperoxidase staining of E11.5 lung buds with rabbit polyclonal anti-NPAS1 antibodies (b–d, e–f) or normal rabbit IgG control antibodies. Lung buds were cultured for 48 h with npas1-sense ODN (b, c, e) or with npas1 AS ODN (d). Bar in lower left hand corner in b = 100 μm, and in c–e = 25 μm. T, trachea; * pleural surface, long arrows indicate NPAS1+ epithelial cells, and short arrow indicates NPAS1+ mesothelial cells. Dotted lines in b demarcate region that is enlarged in c. In c, short red arrowheads indicate strongly NPAS1+ nuclei of parabronchial myofibroblasts just inferior to the tracheal bifurcation. Short black arrow with solid arrowhead in upper right hand corner indicates NPAS1+ neurons and nerve fibers. Long thin arrows flank stretches of NPAS+ myofibroblasts arching along branchopoint, demonstrating apparent nucleolar immunostaining. (f–g) Immunoperoxidase staining of lung tissue from E15.5 (f) and E18.5 (g) embryos with rabbit polyclonal anti-NPAS1 antibodies. (f) At E15.5, there is strongly positive cytoplasmic staining of epithelial cells (long arrows with solid arrowheads), especially for epithelium of larger airways (L = airway lumen), with moderately positive NPAS1 immunostaining of most distal epithelial cells. There is also strong NPAS1+ immunostaining of many nuclei of mesenchymal cells (representative nuclei indicated by short thin arrows). Bar in lower left hand corner = 25 μm; * pleural surface. (g) At E18.5, the only intense npas1 immunostaining is present in nerve fibers and ganglia (short arrow with solid arrowhead, upper right hand corner). NPAS1+ nuclei occur only infrequently in the epithelium (indicated by two long arrows) of airways (L = airway lumen) and primitive alveoli (lower arrow). Bar in lower left hand corner = 25 μm; v, blood vessel.
NPAS1 Immunostaining
We then performed immunohistochemistry to localize NPAS1 protein in paraffin sections of embryonic lung (Figures 1b–1c). E11.5 lung buds were cultured for 48 h with npas1 sense ODN. NPAS1 immunostaining is present in parabronchial mesenchymal cells and a few epithelial cells in the region of branchpoints (Figure 1b, large arrows). At higher magnification, NPAS1 is visualized in nuclei as strong, punctate immunostaining of nucleoli predominantly in mesenchymal cells immediately adjacent to branchpoints (Figure 1c, indicated by long thin arrows). NPAS1 immunostaining of another E11.5 lung bud from the same litter treated with npas1-AS ODN demonstrates abrogation of NPAS1 immunostaining in the parabronchial mesenchyme and the epithelium at the branchpoint (Figure 1d). A lower level of NPAS1 immunostaining remained in the bronchial ganglia (arrow), which is likely to represent relatively higher intracellular levels of NPAS1 in neurons, similar to neuropeptide levels observed using limiting dilution immunoperoxidase analysis (23). As an additional control, a section of the same npas1 sense-treated lung bud shown in Figures 1b and 1c was stained using polyclonal rabbit IgG at the same protein concentration as the affinity-purified rabbit anti-NPAS1. As shown in Figure 1e, this section is devoid of any immunostaining.
We also performed immunostaining for NPAS1 at two later time points during fetal lung development. At E15.5, NPAS1 is strongly detected in nuclei of numerous mesenchymal cells, especially in the vicinity of branchpoints, in the cytoplasm of epithelial cells lining large airways (Figure 1f), and in nerve fibers and ganglia (data not shown). Lower levels of NPAS1 immunostaining are detected in cytoplasm of epithelial cells lining primitive alveoli and in nuclei of less than half of the airway epithelial cells. At E18.5, the only strong NPAS1 immunostaining is present in nerve fibers and ganglia, with weak to moderate staining of nuclei of epithelial cells lining the developing fetal airways (Figure 1g). These temporal and spatial patterns of NPAS1 antigen expression are consistent with a major role for this protein as a regulator of epithelial–mesenchymal interactions during early lung embryogenesis.
NPAS1 and Branching Morphogenesis
We then evaluated whether npas1 or arnt gene expression in embryonic lung might play a functional role in morphogenesis. E11.5 lungs from timed pregnant Swiss-Webster mice were placed in culture with either sense or AS ODN in BGJb media. At this time point, the lung consists of the trachea, primary bronchi, and lung buds corresponding to the five future lobes of the murine lung. The number of lung buds at time zero (E11.5) ranged from five to nine. Littermates were used for all comparisons of sense and AS ODN. Lung buds were photographed at time zero and 48 h later for counting peripheral buds. The percent change from 0 to 48 h was calculated and compared with lungs from littermates treated with the corresponding sense ODN or with media alone. Dose–response studies were performed for npas1 and arnt at 0.5–10 μM final. The greatest effect on lung bud phenotype (see below) was observed at 5–10 μM, so we used 5 μM for all subsequent experiments. QRT-PCR was used to validate reduction of these mRNAs by 5 μM of the AS ODN in E11.5 lung bud cultures (three experiments, three to four lung buds pooled per experiment from each of two litters) compared with the sense controls. Compared with npas1-sense–treated cultures, npas1-AS reduced npas1 mRNA levels by 48.0 ± 0.4% (P < 0.0001). Compared with arnt-sense–treated cultures, arnt-AS reduced arnt mRNA levels by 15 ± 2% (P < 0.0001). Similarly, sim2-AS reduced sim2 mRNA levels by 24 ± 3% (P < 0.0001). Finally, mRNA levels for arnt and sim2 were not significantly altered by npas1-antisense compared with npas1-sense (reduced by 4% and 9%, respectively).
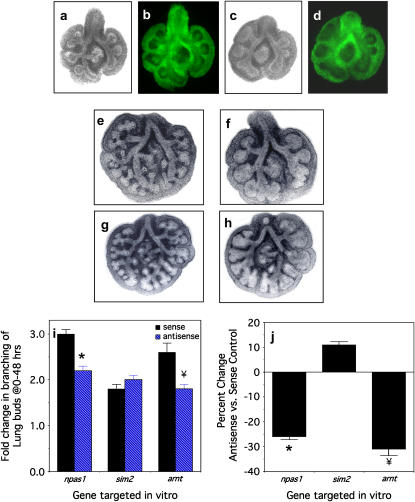
To verify that the ODN were taken up by potential target cells in the lung buds, we added Alex-488–labeled npas1-antisense or -sense ODN to the cultures. After 24 h of incubation, both AS (Figures 2a and 2b) and sense (Figures 2c and 2d) ODN were concentrated predominantly in the mesenchyme (Figures 2b and 2d).
Figure 2.
Branching morphogenesis of murine lung buds is diminished by npas1 or arnt antisense treatment. (a, b) A representative E11.5 lung bud (a, phase contrast; b, fluorescence miscroscopy) treated with fluoresceinated npas1-sense ODN at 5 μM for 24 h demonstrates widespread uptake of the ODN by mesenchymal tissue regions, especially adjacent to the epithelium in the vicinity of branchpoints. (c, d) Similarly, a representative E11.5 lung bud (c, phase contrast; d, fluorescence microscopy) treated with fluoresceinated npas1 AS ODN at 5 μM for 24 h demonstrates widespread uptake of the ODN by mesenchymal tissue regions, especially adjacent to the epithelium in the vicinity of branchpoints. (e) Treatment with npas1-sense at 5 μM for 48 h has no effect on branching morphogenesis of E11.5 lung buds. (f) Treatment of a lung bud from the same litter as in a with npas1 AS leads to decreased numbers of peripheral buds, which are dilated and dysmorphic, ending in rounded sacs. (g) Treatment with arnt-sense at 5 μM for 48 h has no effect on branching morphogenesis. (h) Treatment of lung buds from the same litter as in c with arnt-AS results in decreased numbers of peripheral buds, which are dilated and misshapen, ending in rounded and box-like sacs. (i) The pooled results of the branching assay are given as the percent change in peripheral buds from 0–48 h, with the following numbers of lung buds (3–4 per litter treated with a given ODN: npas1-sense [n = 26] versus npas1 AS [n = 28], * P < 0.0001; arnt-sense [n = 13] versus arnt AS [n = 15], ¥ P < 0.003; and a specificity control, sim2-sense [n = 25] versus sim2 AS [n = 27], P = 0.12). (j) The same data as in (e) is given here as the percent change in branching observed with the AS ODN as compared with the corresponding sense control.
We then used unlabeled ODN for the functional studies. After 48 h, there is no effect of npas1-sense or arnt-sense ODN on branching morphogenesis (Figures 2e and 2g) compared with untreated control lung buds from the same litter (data not shown), whereas npas1-AS and arnt-AS ODN (Figures 2f and 2h) significantly inhibit branching morphogenesis compared with the corresponding sense controls. Compared with the corresponding sense controls, branching morphogenesis of E11.5 lung buds is decreased by 26% by npas1-AS (Figures 2f and 2i), and also reduced 32% by arnt-AS (Figures 2h and 2i). There is no significant difference in branching morphogenesis between lung buds treated with npas1-sense or arnt-sense and media-alone controls from the same litter (data not shown), in contrast to the abnormal morphology of lungs cultured with npas1- or arnt-antisense ODN (Figures 2f and 2h). These data are consistent with heterodimer formation by NPAS1 and Arnt to form a functional complex to regulate transcription, analogous to TRL and Tango in Drosophila (24). Thus, the effect of arnt-AS may be due to decreased NPAS1 function because Arnt is a ubiquitous dimerization partner for many PAS proteins (8, 9). Sim2-antisense did not alter branching morphogenesis compared with the sim2-sense control, whereas sim2-sense alone decreased branching by 29 ± 2% compared with media alone (P = 0.0003). However, sim2-AS did decrease sim2 mRNA levels by 24% compared with sim2-sense, indicating that sim2-AS does have a specific effect on sim2 mRNA levels, in spite of which there is no effect on branching morphogenesis. Thus, effects of antisense ODN are most valid when antisense groups are compared with the corresponding sense control to correct for nonspecific effects of the ODN (25).
We used classical histopathologic methods to compare effects of npas1-AS and arnt-AS on cell proliferation, including both BrdU incorporation and immunostaining for PCNA. Extensive analyses of all sections from multiple experiments demonstrate that cell proliferation is not appreciably altered in npas1- or arnt-AS–treated lungs compared with sense controls (data not shown). Similarly, there is no visible change in apoptosis by npas1-antisense treatment according to either in situ TUNEL analysis with the ApopTag kit or histologic detection of TUNEL-positive apoptotic bodies (data not shown).
Myofibroblasts in NPAS1-AS–Treated Lung Buds
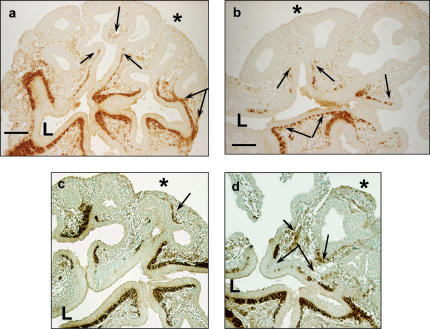
We next used immunohistochemistry to explore cell differentiation in npas1- and arnt-AS–treated lung buds. SMA immunostaining demonstrates a consistently altered distribution of myofibroblasts in E11.5 lung buds cultured with either AS for 2 d. Compared with sense-treated lung buds from littermates (Figures 3a and 3c), we observed reduced extension of myofibroblasts within the subepithelial mesenchyme along the branching epithelial tubes (Figures 3b and 3d). The myofibroblasts in lung buds treated with npas1-AS (Figure 3b) or arnt-AS (Figure 3d) appear as irregular clusters or clumps of cells that are discontinuous and rounded up, extending only about halfway down the primitive bronchial tree (Figure 3d, arrows). In contrast, in sense-treated lungs (Figure 3c, arrows) or lungs cultured without any ODN (data not shown) the myofibroblasts extend evenly and taper off continuously from the proximal airways down to the peripheral buds.
Figure 3.
Immunostaining for SMA in E11.5 lungs treated with npas1- or arnt-antisense. (a) Lungs cultured with npas1 sense ODN have near-continuous SMA staining from the tracheal bifurcation (below “L”), along the conducting airways out to the primitive alveolar buds (as indicated by double arrows). Single arrows indicate the most distal SMA cells. L, proximal airway lumen; * pleural surface; bar in lower left hand corner = 50 μm. (b) Lungs cultured with npas1 AS have discontinuous and/or arrested myofibroblast development occurring about halfway down the developing airway (single arrows). Discontinuous SMA+ immunostaining is indicated by joined double arrows; L, proximal airway lumen; * pleural surface; bar in lower left hand corner = 50 μm. (c) Lungs cultured with arnt-sense also have continuous SMA staining along most of the conducting airways (L, proximal airway lumen) out to near the pleural surface (arrow indicates SMA+ cells near pleural surface, indicated by asterisk). (d) Lungs cultured with arnt AS have discontinuous and/or arrested myofibroblast development along developing airways (arrows). Pleural surface is indicated by an asterisk. Discontinuous SMA+ immunostaining is indicated by double arrows; some SMA+ cells are not clearly associated with airways (small arrowhead). L, proximal airway lumen; * pleural surface; bar in lower left hand corner = 50 μm.
Neuroendocrine Cells in NPAS1-AS–Treated Lung Buds
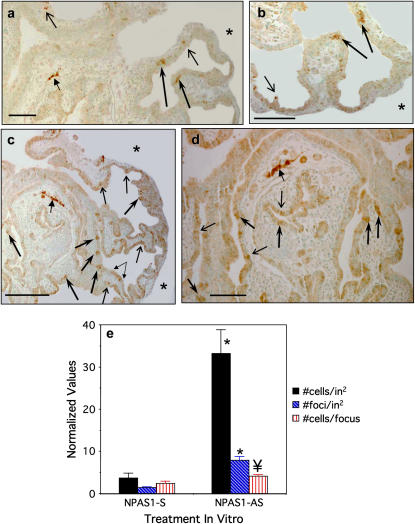
Considering our earlier observations of altered lung branching morphogenesis together with increased numbers of neuroendocrine (NE) cells when Notch signaling is inhibited, we analyzed the expression of an NE cell marker in npas1-AS–treated lung buds (10). We also chose to study NE cells because they are known to be the first epithelial cell to differentiate in ontogeny and can be detected after about one week of culture of E11.5 lung buds (26). After 6 d in culture, occasional NE cells can be visualized in npas1-sense–treated controls by immunostaining for the early neural/NE cytoplasmic marker PGP9.5 (Figures 4a and 4b). Other NE markers such as calcitonin gene-related peptide are not yet detectable (data not shown). These NE cells are distributed both proximally and distally in the primitive lung buds, extending from the mainstem bronchi down at least three airway generations. Lung buds treated with media alone without ODN for 6 d also had infrequent NE cells (data not shown). In contrast, lung buds treated with npas1-AS for 6 d (Figures 4c and 4d) have markedly increased numbers of NE cells, occurring both as single cells (thin arrows) and as clusters within the primitive airway epithelium (thick arrows). Occasionally linear arrays of NE cells are seen (double arrow in Figure 4c). In npas1-AS–treated lung buds, the NE cells are distributed both proximally and distally, beginning in the mainstem bronchi, and throughout the epithelium for at least three airway generations. Quantification of relative numbers of NE cells in these lung buds normalized for the tissue area of each lung bud is shown in Figure 4e. In lung buds treated for 6 d with npas1-AS ODN, there is a highly significant increase in both the number of PGP+ foci (5.6-fold increased) and the total number of NE cells (9-fold increased) per in2 area of lung bud tissue (P < 0.0001). There is also a significant increase in the number of NE cells per focus (1.7-fold increased, P < 0.012).
Figure 4.
PGP9.5 Immunostaining in E11.5 lungs treated with npas1- or arnt-antisense. (a, b) Immunostaining of lung buds cultured for 6 d with npas1-sense oligos have occasional clusters of PGP9.5-positive NE cells scattered throughout the airway epithelium (long dark arrows) and also a few isolated NE cells (short thin arrows). As an internal positive control, a PGP9.5-positive nerve fiber is visible in the mid-left field of a (solid arrowhead). Bars in lower left hand corner = 50 μm. * Pleural surface. (c, d) Lungs cultured with npas1 AS for 6 d have a marked increase in PGP9.5-positive NE cell clusters (solid arrows), as well as a modest increase in isolated NE cells (thin arrows). Small linked arrows in lower right hand corner of c indicate a stretch of linear NE cell hyperplasia. The internal positive control is a PGP9.5-positive nerve fiber (arrowhead, upper left hand corner of c, in which asterisk indicates pleural surface). Bars in lower left hand corner = 100 μm (c) and 50 μm (d). (e) Morphometric analysis for relative numbers of PGP9.5+ NE cells in E11.5 lung buds treated with npas1-sense or -AS ODN in culture for 6 d. We determined the number of PGP+ cell foci (including both isolated cells and clusters of ⩾ 2 cells) and the total number of PGP+ cells, both of which were normalized for the area of lung tissue present using Scion Image 1.62 software (Scion Corp., Frederick, MD) and density slice analysis. The number of PGP+ cells per focus was also calculated. Data shown represent the mean values ± SE for complete cross-sections of n = 9 (sense-treated lung buds) and n = 14 (AS-treated lung buds), representing pooled data from four experiments including matched controls from the same litters. * P < 0.0001 compared with npas1 sense control; ¥ P < 0.012 compared with sense control.
mRNA Microarray Analyses
To begin to investigate molecular mechanisms underlying the observed morphologic changes, we performed expression profiling of lung buds treated with npas1-AS versus -sense controls from the same litters. RNA was extracted from pooled lung samples from five separate experiments (three to six lung buds per experiment) after 48 h of culture. Microarray analysis was performed by probing five pairs of Affymetrix 430A-2.0 gene array chips (Affymetrix, Union City, CA) using cDNA prepared from npas1-AS– versus npas1-sense–treated lung buds. Detailed protocols are given elsewhere (27).
The cumulative data were analyzed by excluding all samples with an absent signal in either AS- or sense-treated mRNA samples. The remaining genes were sorted and selected for P < 0.05 and fold-change > 3-fold. We identified a total of 157 genes consistently down-regulated and 11 genes consistently up-regulated in all five experiments (listed in Table E3). Of these genes, 9 are duplicates and 62 are unidentified genes (24 RIKENs, 20 unknowns, 1 open reading frame, and 17 ESTs). Excluding these genes results in 93 genes down-regulated by 1/3 or less by npas1-AS and 5 genes up-regulated by 3-fold or more by npas1-AS. None of the up-regulated genes appear to be of functional significance. At least 13 of the genes down-regulated by npas1-AS have known functions in regulating cell shape, cell adhesion, cell migration, extracellular matrix formation, and/or smooth muscle cell differentiation (Table 1). At least 4 of these genes plus 12 additional genes are known to play a role in normal development and/or angiogenesis, including 5 transcription factors (Table 1) (also listed in Tables E3 and E4). Other down-regulated genes are components of signal transduction cascades, many of which are involved in G-protein signaling, which can regulate cell migration. Finally, some of the genes down-regulated by npas1-AS are likely to play a role in lung development, including sonic hedgehog (Shh) signaling molecules (hhip, Hoxb4, Tbx5, dachshund, IGF-I).
TABLE 1.
GENES DOWN-REGULATED IN LUNG BUDS BY NPAS1-ANTISENSE
| Cell migration/adhesion/muscle development |
| Actin, alpha 2, smooth muscle, aorta* |
| Actin, gamma 2, smooth muscle, enteric |
| Angiopoietin 2 (inhibits angiogenesis)* |
| Claudin 11 |
| Elfin (PDZ and LIM domain 1) |
| Junction cell adhesion molecule 3 |
| Protocadherin beta 9 (Pcdhb9) |
| RHAMM (receptor for hyaluronic acid)* |
| Myocardin |
| Rab-2 (GTPase)\rho GTPase-activating protein 6 |
| Transforming growth factor-beta induced, 68 kD |
| Transgelin |
| Tropomyosin 2, beta* |
| Cell differentiation/morphogenesis |
| Transcription Factors |
| Forkhead box C1 (Foxc1) |
| Homeobox B4 (Hoxb4) |
| Recombining binding protein suppressor of hairless (Rbpsuh) |
| Transducin-like enhancer of split 1 homolog (Tle1) |
| T-box transcription factor (Tbx5) |
| Other regulatory molecules |
| Adenylate cyclase 3 |
| Endothelial differentiation, G-protein–coupled receptor 2* |
| Faciogenital dysplasia homolog |
| Fibrosin |
| Hedgehog-interacting protein (hedgehog antagonist) |
| Inhibitor of kappaB kinase gamma |
| Syndecan 2 |
Role in angiogenesis.
Some genes down-regulated by npas1-AS are known regulators of CNS development, including two neurogenic genes (RBPsuh, suppressor-of-hairless, and Tle, transducin-like enhancer-of-split), which function in Notch-related lateral inhibition of neural/NE differentiation (28). This gene family also plays a major downstream role in tracheal development in Drosophila (9). Decreased expression of neurogenic genes would provide a mechanistic basis for increased pulmonary NE cells in npas1-AS–treated lungs.
To validate expression levels of several genes that would likely contribute to the altered morphogenesis that we observed in the lung buds, we performed QRT-PCR. The results of three pooled experiments are given in Figure 5. Npas1 mRNA is reduced ∼ 50%, consistent with earlier experiments. Both RBPsuh and Tle are decreased ∼ 20%, which seems modest but is statistically significant. Diminished expression of both of these genes could function synergistically, because they are known to act in the same signaling pathway, leading to the observed increase in NE cells after 6 d in culture. Furthermore, notch-1 AS treatment has been demonstrated to induce phenotypic changes in chick retina with as little as 20% reduction in notch-1 mRNA levels (29). Last, but not least, RBP and other neurogenic genes in the Notch signaling pathway also play a major role in the determination of smooth muscle differentiation (30, 31), consistent with pleiotropic effects of decreased RBPsuh contributing to both increased NE cells and decreased smooth muscle development in E11.5 lung buds.
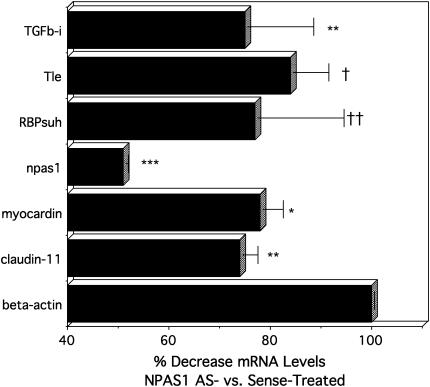
Figure 5.
QRT-PCR validation of differential gene expression in E11.5 lung buds treated with NPAS1-antisense versus -sense ODN. The results given are the pooled data from three separate experiments using three independent RNA preparations from E11.5 lung buds treated with ODN for 48 h. Results are presented as mean percentage change ± SE, comparing npas1-AS–treated lungs versus npas1-sense–treated lungs. For comparison, β-actin is defined as 100% for npas1-AS relative to sense. Significance levels are: *** P < 0.0001; ** P < 0.005; * P < 0.01; †† < 0.02; † P < 0.05.
Gene expression for TGF-β-induced 68 kD, also termed “β-IG-H3” are reduced ∼ 30% (P < 0.005). BetaIG-H3 is likely to play a role in lung development, being expressed at high levels in distal lung during organogenesis (32). It is present in the extracellular matrix, cytoplasm, and nuclei of smooth muscle cells in human airways and pulmonary vasculature, especially in the septal tips of alveolar ducts and alveoli, suggesting that it may have a morphogenetic role (33).
Claudin-11, also significantly decreased by npas1-AS with P < 0.005, is a transmembrane tight-junction protein that is homologous to Drosophila Megatrachea, which plays an important role in regulating morphogenesis of the Drosophila tracheal system (34). Claudin-11 deficiency could also lead to the distal airspace dilatation that we observe in npas1-AS–treated lung buds. Claudin-3 is expressed in chick lung buds during development (35). Claudin-5 localizes to junctions between cardiomyocytes and its deficiency is associated with dilated cardiomyopathy (36), suggesting that decreased claudin-11 could be linked to the abnormal shape and lack of continuity between myofibroblasts along airways of npas1-AS–treated lung buds.
Finally, down-regulation of myocardin gene expression (P < 0.01) could be consistent with impaired development of smooth muscle along developing airways (37). Myocardin is a transcription factor that has been shown to be necessary and sufficient for smooth muscle cell differentiation (37, 38).
Conclusions
In summary, we have demonstrated that npas1 and arnt are expressed and functional in murine embryonic lung: knocking down npas1 or arnt mRNA levels in lung bud cultures inhibits lung branching and myofibroblast development, analogous to decreased cell migration in Trl-null Drosophila. Furthermore, npas1-AS–treated lungs have down-regulated expression of numerous developmentally important genes. Two prominent clusters of inter-related genes that are down-regulated by npas1-AS are: first, genes controlling cell migration, adhesion, and/or smooth muscle development; and second, genes regulating neurogenesis. These observations provide a mechanistic basis for inhibition of branching and smooth muscle development and increased numbers of NE cells.
Myofibroblast migration is recognized to be a major factor in normal lung morphogenesis, especially during alveolarization (39). Similarly in Drosophila TRL functions by regulating cell migration during formation of the primary tracheal tube and also during branching morphogenesis (6, 9). Previously we demonstrated that notch-1-AS–treated lung buds have increased branching morphogenesis and increased numbers of NE cells (10). In Drosophila, Notch is implicated as a negative regulator of tracheal development downstream from TRL (9). The cumulative direct and circumstantial evidence suggests that npas1 may be one functional mammalian homolog of the Drosophila trl gene. Despite high sequence conservation, in the CNS several other PAS proteins have been found to be biologically essential and functionally nonredundant (8). However, functional redundancy is common in developmental/biological processes that are essential for survival. We propose that functional redundancy of multiple signaling pathways activated by npas1 could provide a selective advantage to preserve normal lung structure and function that are crucial for postnatal survival. Thus, our short-term in vitro studies are not incompatible with the apparent lack of a pulmonary phenotype in NPAS1 KO mice. In conclusion, the cumulative evidence indicates that NPAS1 can function as a proximal regulator of lung organogenesis, consistent with NPAS1 being a candidate mammalian homolog of TRL, the primary regulator of respiratory system development in Drosophila.
Supplementary Material
Acknowledgments
The authors thank Dr. Vance Morgan, Linette Mookanamparambil, and Deepti Anand in the Harvard Partners Center for Genetics and Genomics for carrying out the microarray analyses. The authors acknowledge the technical assistance of Fan Zhang.
This work was supported by NIH grant #2RO1 HL44984 (M.E.S.) and NRSA training grant (B.M.L.).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2006-0314OC on November 16, 2006
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol 1999;209:60–71. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Izvolsky KI, Qian J, Cardoso WV. Identification of FGF10 targets in the embryonic lung epithelium during bud morphogenesis. J Biol Chem 2005;280:4834–4841. [DOI] [PubMed] [Google Scholar]
- 3.Weaver M, Batts L, Hogan BL. Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev Biol 2003;258:169–184. [DOI] [PubMed] [Google Scholar]
- 4.van Tuyl M, Post M. From fruitflies to mammals: mechanisms of signalling via the Sonic hedgehog pathway in lung development. Respir Res 2000;1:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manning G, Krasnow MA. in Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; 1993. pp. 609–685.
- 6.Wilk R, Weizman I, Shilo B-Z. Trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev 1996;10:93–102. [DOI] [PubMed] [Google Scholar]
- 7.Affolter M, Bellusci S, Itoh N, Shilo B, Thiery JP, Werb Z. Tube or not tube: remodeling epithelial tissues by branching morphogenesis. Dev Cell 2003;4:11–18. [DOI] [PubMed] [Google Scholar]
- 8.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol 2004;36:189–204. [DOI] [PubMed] [Google Scholar]
- 9.Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science 1999;284:1635–1639. (Review). [DOI] [PubMed] [Google Scholar]
- 10.Kong Y, Glickman J, Subramaniam M, Shasafaei A, Allamneni KP, Aster JC, Sklar J, Sunday ME. Functional diversity of notch family genes in fetal lung development. Am J Physiol Lung Cell Mol Physiol 2004;286:L1075–L1083. [DOI] [PubMed] [Google Scholar]
- 11.Brunskill EW, Witte DP, Shreiner AB, Potter SS. Characterization of npas3, a novel basic helix-loop-helix PAS gene expressed in the developing mouse nervous system. Mech Dev 1999;88:237–241. [DOI] [PubMed] [Google Scholar]
- 12.Zhou YD, Barnard M, Tian H, Li X, Ring HZ, Francke U, Shelton J, Richardson J, Russell DW, McKnight SL. Molecular characterization of two mammalian bHLH-PAS domain proteins selectively expressed in the central nervous system. Proc Natl Acad Sci USA 1997;94:713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci USA 2004;101:13648–13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunskill EW, Ehrman LA, Williams MT, Klanke J, Hammer D, Schaefer TL, Sah R, Dorn GW 2nd, Potter SS, Vorhees CV. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur J Neurosci 2005;22:1265–1276. [DOI] [PubMed] [Google Scholar]
- 15.Garcia JA, Zhang D, Estill SJ, Michnoff C, Rutter J, Reich M, Scott K, Diaz-Arrastia R, McKnight SL. Impaired cued and contextual memory in NPAS2-deficient mice. Science 2000;288:2226–2230. [DOI] [PubMed] [Google Scholar]
- 16.Herradon G, Ezquerra L, Nguyen T, Silos-Santiago I, Deuel TF. Midkine regulates pleiotrophin organ-specific gene expression: evidence for transcriptional regulation and functional redundancy within the pleiotrophin/midkine developmental gene family. Biochem Biophys Res Commun 2005;333:714–721. [DOI] [PubMed] [Google Scholar]
- 17.Iacobas DA, Urban M, Iacobas S, Spray DC. Transcriptomic characterization of four classes of cell-cell/cell-matrix genes in brains and hearts of wild type and connexin43 null mice. Rom J Physiol 2002. –2003;39–40:91–116. [PubMed] [Google Scholar]
- 18.Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc Natl Acad Sci USA 2000;97:8386–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asikainen TM, Waleh NS, Schneider BK, Clyman RI, White CW. Enhancement of angiogenic effectors through hypoxia-inducible factor in preterm primate lung in vivo. Am J Physiol Lung Cell Mol Physiol 2006;291:L588–L595. [DOI] [PubMed] [Google Scholar]
- 20.Shan L, Levesque B, Kong Y, Sunday ME. Genes encoding neuronal PAS domain proteins (NPAS1 & 3) and aryl hydrocarbon receptor nuclear translocator (ARNT) are expressed in developing mouse lung. Am J Respir Crit Care Med 2004;169:A662. [Google Scholar]
- 21.Levesque BM, Kong Y, Shan L, Sunday ME. PAS domain transcription factors (TFs) regulate early lung morphogenesis. Am J Respir Crit Care Med 2003;167:A381. [Google Scholar]
- 22.Goshu E, Jin H, Fasnacht R, Sepenski M, Michaud JL, Fan CM. Sim2 mutants have developmental defects not overlapping with those of Sim1 mutants. Mol Cell Biol 2002;22:4147–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Springall DR, Collina G, Barer G, Suggett AJ, Bee D, Polak JM. Increased intracellular levels of calcitonin gene-related peptide-like immunoreactivity in pulmonary endocrine cells of hypoxic rats. J Pathol 1988;155:259–267. [DOI] [PubMed] [Google Scholar]
- 24.Sonnenfeld MJ, Delvecchio C, Sun X. Analysis of the transcriptional activation domain of the Drosophila Tango bHLH-PAS transcription factor. Dev Genes Evol 2005;215:221–229. [DOI] [PubMed] [Google Scholar]
- 25.Scherer LJ, Rossi JJ. Approaches for the sequence-specific knockdown of mRNA. Nat Biotechnol 2003;21:1457–1465. [DOI] [PubMed] [Google Scholar]
- 26.Ten Have-Opbroek AAW. Lung development in the mouse embryo. Exp Lung Res 1991;17:111–130. [DOI] [PubMed] [Google Scholar]
- 27.Sonis S, Haddad R, Posner M, Watkins B, Fey E, Morgan TV, Mookanamparambil L, Ramoni M. Gene expression changes in peripheral blood cells provide insight into the biological mechanisms associated with regimen-related toxicities in patients being treated for head and neck cancers. Oral Oncol 2007;43:289–300. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Baker NE. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol 2001;11:330–338. [DOI] [PubMed] [Google Scholar]
- 29.Austin CP, Feldman DE, Ida JA Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development 1995;121:3637–3650. [DOI] [PubMed] [Google Scholar]
- 30.Doi H, Iso T, Sato H, Yamazaki M, Matsui H, Tanaka T, Manabe I, Arai M, Nagai R, Kurabayashi M. Jagged1-selective notch signaling induces smooth muscle differentiation via RBP-Jkappa dependent pathway. J Biol Chem 2006;281:28555–28564. [DOI] [PubMed] [Google Scholar]
- 31.Gao X, Chandra T, Gratton MO, Quelo I, Prud'homme J, Stifani S, St-Arnaud R. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J Cell Biol 2001;154:1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Qian J, Izvolsky KI, Cardoso WV. Global analysis of genes differentially expressed in branching and non-branching regions of the mouse embryonic lung. Dev Biol 2004;273:418–435. [DOI] [PubMed] [Google Scholar]
- 33.Billings PC, Herrick DJ, Howard PS, Kucich U, Engelsberg BN, Rosenbloom J. Expression of betaig-h3 by human bronchial smooth muscle cells: localization To the extracellular matrix and nucleus. Am J Respir Cell Mol Biol 2000;22:352–359. [DOI] [PubMed] [Google Scholar]
- 34.Behr M, Riedel D, Schuh R, Fujita M, Itoh M, Shibata M, Taira S, Taira M. The claudin-like megatrachea is essential in septate junctions for the epithelial barrier function in Drosophila. Dev Cell 2003;5:611–620. [DOI] [PubMed] [Google Scholar]
- 35.Haworth KE, El-Hanfy A, Prayag S, Healy C, Dietrich S, Sharpe P. Expression of Claudin-3 during chick development. Gene Expr Patterns 2005;6:40–44. [DOI] [PubMed] [Google Scholar]
- 36.Sanford JL, Edwards JD, Mays TA, Gong B, Merriam AP, Rafael-Fortnoy JA. Claudin-5 localizes to the lateral membranes of cardiomyocytes and is altered in utrophin/dystrophin-deficient cardiomyopathic mice. J Mol Cell Cardiol 2005;38:323–332. [DOI] [PubMed] [Google Scholar]
- 37.Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev 2004;14:558–566. [DOI] [PubMed] [Google Scholar]
- 38.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev 2006;20:1545–1556. [DOI] [PubMed] [Google Scholar]
- 39.Brody JS, Vaccaro C. Postnatal formation of alveoli: interstitial events and physiologic consequences. Fed Proc 1979;38:215–223. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.