Abstract
Staphylococcus aureus lipoteichoic acid (LTA) is composed of a linear 1,3-linked polyglycerolphosphate chain and is tethered to the bacterial membrane by a glycolipid (diglucosyl-diacylglycerol [Glc2-DAG]). Glc2-DAG is synthesized in the bacterial cytoplasm by YpfP, a processive enzyme that transfers glucose to diacylglycerol (DAG), using UDP-glucose as its substrate. Here we present evidence that the S. aureus α-phosphoglucomutase (PgcA) and UTP:α-glucose 1-phosphate uridyltransferase (GtaB) homologs are required for the synthesis of Glc2-DAG. LtaA (lipoteichoic acid protein A), a predicted membrane permease whose structural gene is located in an operon with ypfP, is not involved in Glc2-DAG synthesis but is required for synthesis of glycolipid-anchored LTA. Our data suggest a model in which LtaA facilitates the transport of Glc2-DAG from the inner (cytoplasmic) leaflet to the outer leaflet of the plasma membrane, delivering Glc2-DAG as a substrate for LTA synthesis, thereby generating glycolipid-anchored LTA. Glycolipid anchoring of LTA appears to play an important role during infection, as S. aureus variants lacking ltaA display defects in the pathogenesis of animal infections.
Lipoteichoic acid (LTA) is an abundant secondary wall polymer in the cell wall envelope of gram-positive bacteria (11, 13, 27). LTA plays an important role during host infection, as it is thought to be perceived by receptors on immune cells that trigger innate responses in an effort to defend host tissues from invading microbes (34). Other proposed functions of LTA include Mg2+ ion scavenging and proper targeting of autolysins to the bacterial envelope. Physiological functions of autolysins in degradation of cell wall envelopes or separation of dividing cells are essential for bacterial growth (6, 16, 31).
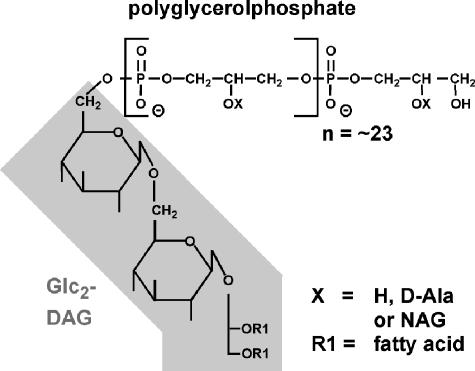
The chemical structure of LTA has been determined for several different gram-positive bacteria (13). Staphylococcus aureus LTA is composed of 1,3-linked polyglycerolphosphate chains linked to β-gentiobiosyldiacylglycerol (diglucosyl-diacylglycerol [Glc2-DAG]), which anchors LTA in the bacterial membrane (8). Under laboratory growth conditions, 75 to 80% of LTA glycerolphosphate moieties have d-alanine ester substitutions at position 2 (15), and some staphylococcal strains have N-acetylglucosamine substitutions at the same position (43) (Fig. 1). The polyglycerolphosphate structure of LTA is found in many other gram-positive bacteria, including Bacillus subtilis, Bacillus licheniformis, Bacillus cereus, Bacillus anthracis, Enterococcus faecalis, Listeria spp., and group A and group B streptococci (11, 14).
FIG. 1.
Structure of S. aureus lipoteichoic acid. S. aureus LTA is composed of linear polyglycerolphosphate chains (average length, 23 repeating units), which are linked to a membrane anchor composed of Glc2-DAG (shaded box). Position 2 (indicated by X) of the repeating glycerolphosphate subunits can be unsubstituted (hydrogen), esterified with d-alanyl, or linked to N-acetylglucosamine (NAG). The diagram was adapted from previously published schemes (12, 35).
YpfP, a processive glycosyltransferase, is required for glycolipid synthesis in B. subtilis and S. aureus (24, 25, 27). Expression of B. subtilis ypfP in Escherichia coli, an organism that does not synthesize glycolipids, leads to accumulation of several glycolipids and phosphoglycolipids (24, 25). Inactivation of ypfP in S. aureus abrogates all glycolipid synthesis and leads to morphological alterations, including an increase in cell size and aberrant cell shapes (27). Interestingly, LTA synthesis is not abolished in a ypfP mutant strain, and LTA with wild-type chain length is produced (27). However, LTA is anchored to the membrane via diacylglycerol (DAG) in the mutant, and increased amounts of LTA are found in the culture supernatant (27). Similarly, a defect in glycolipid synthesis in group B Streptococcus leads to shedding of LTA into the medium, and mutant bacteria have defects in invasion of eukaryotic cells and virulence (7).
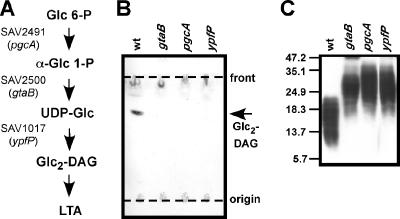
YpfP successively transfers glucose onto DAG using UDP-glucose as its substrate (24, 25). In B. subtilis, UDP-glucose is synthesized by conversion of glucose-6-phosphate to α-glucose-1-phosphate, a reaction catalyzed by α-phosphoglucomutase (PgcA; formerly known as GtaC and GtaE) (32). GtaB (UTP:α-glucose-1-phosphate uridyltransferase) synthesizes UDP-glucose from α-glucose-1-phosphate and UTP (41, 46) (Fig. 2A). Similar to ypfP mutants, B. subtilis or B. licheniformis mutants unable to produce UDP-glucose are defective in the synthesis of glycolipids and glycolipid-anchored LTA (4, 32).
FIG. 2.
(A) LTA glycolipid anchor synthesis in S. aureus. Glucose-6-phosphate (Glc 6-P) is converted by α-phosphoglucomutase (PgcA, encoded by SAV2941) to α-glucose-1-phospate (α-Glc 1-P), which is then activated by UTP:α-glucose-1-phosphate uridyltransferase (GtaB, encoded by SAV2500) to generate UDP-glucose (UDP-Glc). UDP-Glc and diacylglycerol (DAG) serve as substrates for YpfP, a processive glycosyltransferase that generates diglucosyl-diacylglycerol (Glc2-DAG), which, after transfer of linear polyglycerolphosphate, functions as the membrane anchor for LTA. (B) TLC of glycolipids from S. aureus. Samples (300 μg) of membrane lipids isolated from S. aureus strains ANG361 (wild type) (wt), ANG372 (gtaB), ANG371 (pgcA), and ANG370 (ypfP) were separated by TLC, and glycolipids were visualized with α-naphthol/sulfuric acid. The positions of the origin and TLC solvent front are indicated by dashed lines. (C) Immunoblot analysis of LTA extracted from S. aureus. LTA was extracted from S. aureus, separated by 15% SDS—PAGE, and detected with a polyglycerolphosphate-specific antibody. The positions of protein molecular mass markers (in kDa) are indicated on the left.
Using bioinformatic tools, we identified S. aureus homologs of B. subtilis pgcA and gtaB. Inactivation of pgcA or gtaB eliminated the synthesis of membrane glycolipids in S. aureus. Similar to an S. aureus ypfP mutant, variants lacking pgcA or gtaB had the ability to generate LTA. A new gene, ltaA (lipoteichoic acid gene A) that is located in the same operon and is immediately adjacent to ypfP was identified. While ltaA is dispensable for the synthesis of membrane glycolipids, it is required for the efficient synthesis of LTA with a glycolipid anchor.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strains were grown in Luria-Bertani medium, and S. aureus strains were grown in tryptic soy broth (TSB) at 37°C. All bacterial strains used in this study are listed in Table 1. S. aureus strain SEJ1 is a derivative of strain RN4220 (29) with an in-frame deletion in spa (encoding protein A). S. aureus strains harboring mariner transposon insertions in SAV1016 (ltaA), SAV1017 (ypfP), SAV2491 (pgcA), and SAV2500 (gtaB) were grown in TSB supplemented with 10 μg/ml erythromycin. Ampicillin (100 μg/ml) and chloramphenicol (10 μg/ml) were used for selection of plasmid pCL55 (33) and its derivatives in E. coli and S. aureus, respectively. Gene expression from the tetracycline-inducible promoter was induced by addition of 200 ng/ml anhydrotetracycline.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant features | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| XL1-Blue | Stratagene | |
| XL1-Blue/pYJ335 | E. coli/S. aureus shuttle vector containing a tetracycline-inducible promoter | 23 |
| ANG243 | pCL55 in XL1-Blue; S. aureus single-site integration vector | 33 |
| ANG284 | pitet in XL1-Blue; pCL55 containing a tetracycline-inducible promoter | This study |
| ANG373 | pCL55-ypfP in XL1-Blue; expression of YpfP (SAV1017) under native promoter control | This study |
| ANG374 | pCL55-ypfP/ltaA in XL1-Blue; expression of YpfP (SAV1017) and LtaA (SAV1016) under native promoter control | This study |
| ANG375 | pitet-ltaA in XL1-Blue; expression of LtaA (SAV1016) under tetracycline-inducible promoter control | This study |
| Staphylococcus aureus strains | ||
| Newman | Human clinical isolate | 9 |
| ΦΝΞ171-39 | Newman with transposon insertion in SAV1017 (ypfP) | 2 |
| ΦΝΞ177-2 | Newman with transposon insertion in SAV2491 (pgcA) | 2 |
| ΦΝΞ1952 | Newman with transposon insertion in SAV2500 (gtaB) | 2 |
| ANG460 | Newman with transposon insertion in SAV1016 (ltaA); transduced from strain ANG359 | This study |
| RN4220 | Transformable laboratory strain | 29 |
| SEJ1 | RN4220Δspa; in-frame deletion in spa coding for protein A | Taeok Bae |
| ANG359 | SEJ1 with transposon insertion in SAV1016 (ltaA); transduced | This study |
| ANG361 | SEJ1 with transposon insertion in COL2559; transduced; wild type for LTA expression used as control strain | This study |
| ANG370 | SEJ1 with transposon insertion in SAV1017 (ypfP); transduced from strain ΦΝΞ171-39 | This study |
| ANG371 | SEJ1 with transposon insertion in SAV2491 (pgcA); transduced from strain ΦΝΞ177-2 | This study |
| ANG372 | SEJ1 with transposon insertion in SAV2500 (gtaB); transduced from strain ΦΝΞ1952 | This study |
| ANG391 | pitet-ltaA in strain ANG359 (ltaA [SAV1016] mutant) | This study |
| ANG393 | pitet in strain ANG359 (ltaA [SAV1016] mutant) | This study |
| ANG395 | pitet in strain ANG361 (wild type for LTA) | This study |
Transposon insertion and phage transduction.
Transposon insertion sites were determined by inverse PCR and DNA sequencing, as previously described (2). Strains ANG370 (transposon insertion in SAV1017 [ypfP]), ANG371 (insertion in SAV2941 [pgcA]), and ANG372 (insertion in SAV2500 [gtaB]) were generated by transducing transposons from bursa aurealis library mutants ΦΝΞ171-39, ΦΝΞ177-2, and ΦΝΞ1952 (2) into strain SEJ1. In a separate study, mariner transposon mutagenesis was performed with S. aureus SEJ1. ANG361 (control strain with transposon insertion at an irrelevant site) and ANG359 (transposon insertion in SAV1016 [ltaA]) were obtained by moving transposon insertions from two of these library strains into a fresh SEJ1 background strain. Strain ANG460 was obtained by moving a transposon from strain ANG359 (SAV1016 [ltaA]) into S. aureus strain Newman. Strain ANG460 and parental strain Newman were used for virulence studies.
Lipoteichoic acid immunoblotting.
For sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blot analysis of cell-associated LTA, 1 ml of a staphylococcal overnight culture was mixed with 0.5 ml of 0.1-mm glass beads, and the bacteria were lysed by 45 min of vortexing in the cold. The glass beads and cell debris were sedimented by centrifugation at 200 × g for 1 min, and 0.5 ml of supernatant was transferred to a new tube. Staphylococcal membranes and LTA were sedimented by centrifugation at 16,000 × g for 10 to 15 min and suspended in 100 μl of sample buffer containing 2% SDS. Where indicated below, samples were normalized based on the optical density at 600 nm (OD600); that is, samples from a culture with an OD600 of 6 were suspended in 90 μl sample buffer. To determine the amount of LTA shed into the culture medium, samples were prepared as follows. Bacteria from a 500-μl culture were pelleted by centrifugation at 16,000 × g for 5 min. One hundred microliters of culture supernatant was removed and mixed 1:1 with sample buffer. Samples were boiled for 20 to 30 min, insoluble material removed by centrifugation at 16,000 × g for 5 min, and samples were subjected to 15% SDS-PAGE followed by electrotransfer to polyvinylidene difluoride membranes. Supernatant LTA samples were normalized based on OD600; that is, 10 μl of a culture with an OD600 of 5 was loaded. LTA (polyglycerolphosphate)-specific primary antibody (clone 55; HyCult Biotechnology) and horseradish peroxidase-linked anti-mouse secondary antibody (Cell Signaling) were used at dilutions of 1:2,500 and 1:5,000. Immunoreactive LTA species were detected by enhanced chemiluminescence.
Plasmid and strain construction.
S. aureus RN4220 (29) chromosomal DNA was used as a template for PCRs. For expression of SAV1017 (ypfP) and SAV1017/SAV1016 (ypfP/ltaA) from the native promoter, plasmids pCL55-ypfP and pCL55-ypfP/ltaA were constructed. Primers 5-BamHI-P-SAV1017 (CGGGATCCGCTCCTTTTTCTACAATATGTTTATTATACACG) and 3-KpnI-SAV1017 (GGGGTACCTTATTTAACGAAGAATCTTGCATATAAAGGAACC) and primers 5-BamHI-P-SAV1017 and 3-KpnI-SAV1016 (GGGGTACCTTACTTAGCTTTTTCTCTATTTGCTATAAAGTAGC) were used to amplify SAV1017 (ypfP) and SAV1017/SAV1016 (ypfP/ltaA) DNA, respectively. PCR products were cut with BamHI/KpnI and ligated with chromosomal integration vector pCL55 (33) cut with the same enzymes. For tetracycline-inducible gene expression, pitet was constructed. Primer 5-KpnI-tet (GGGGTACCTTGGTTACCGTGAAGTTACCATCACGG) and the 5′-phosphorylated and restriction site-containing primer P-revSacIIPmeIBglIIAvrII-tet P-(CCGCGGGTTTAAACAGATCTCCTAGGTCATTTGATATGCCTCCGAATTCG) were used to amplify the inducible tetracycline region from plasmid pYJ335 (23). The resulting PCR product was cut with KpnI and ligated with pCL55 that had been digested with KpnI/SmaI to generate pitet. pitet-ltaA was constructed for tetracycline-inducible expression of SAV1016 (ltaA). Primers 5-AvrII-SAV1016 (CCGCCTAGGCATCACAACCACAAGAGATTTATGGAAAGGTTCC) and 3-SacII-SAV1016 (TCCCCGCGGTTACTTAGCTTTTTCTCTATTTGCTATAAAGTAGC) were used to amplify SAV1016. The PCR product was digested with AvrII/SacII and ligated with pitet that had been cut with the same enzymes. All plasmids were cloned in E. coli strain XL1-Blue, and the DNA sequences of inserts were verified by fluorescent automated sequencing at the DNA sequencing facility of the University of Chicago Cancer Research Center. Plasmids were then integrated into the geh locus (lipase gene) in the S. aureus chromosome.
Membrane lipid isolation.
Lipids from S. aureus and E. coli strains were extracted by using a modified Bligh-Dyer method (26). For isolation of S. aureus membrane lipids, 0.1 or 1 liter of TSB was inoculated with 1 or 10 ml of an overnight culture, and the resulting culture was grown at 37°C until the OD600 was ∼3.5. Cultures were chilled on ice for 30 to 60 min, and staphylococci were collected by centrifugation. Bacteria were washed with 0.1 M sodium acetate (pH 4.7) or 0.1 M sodium citrate (pH 4.7) and lysed by shearing with 0.1-mm glass beads. For 0.1-liter cultures, bacterial suspensions were placed in 2-ml Fast Prep tubes containing 0.1-mm glass beads (∼0.5 ml) and lysed with a Fast-Prep machine (Q-BIOgene) by shaking the tubes three times for 45 s at setting 6. The glass beads were sedimented by centrifugation at 200 × g for 1 min, and the supernatant containing bacterial lysate was transferred to new tubes. Lysed bacteria were sedimented by centrifugation at 16,000 × g for 5 min, and lipids were extracted. For large cultures, 40 ml of a washed bacterial suspension was mixed with an equal volume of glass beads; the bacteria were lysed with a bead beater (Biospec Products, Inc.) by shaking samples three times for 2 min at 4°C. Bacteria were chilled between each run for 5 min on ice. The glass beads were sedimented by centrifugation for 1 min at 200 × g. The membranes were removed with the supernatant, and the bacterial debris was sedimented by centrifugation at 12,000 × g for 20 min. The pellets were washed with 40 ml of 0.1 M sodium acetate (pH 4.7) or 0.1 M sodium citrate (pH 4.7), the wet weight was determined, and samples were stored frozen at −20°C. For lipid extraction, frozen pellets were suspended (0.4 g/ml) in 0.1 M sodium acetate (pH 4.7) or 0.1 M sodium citrate (pH 4.7). Chloroform and methanol were added to obtain a final methanol/chloroform/buffer ratio of 2:1:0.8. Lipids were extracted for 2 h at room temperature with vortexing. Insoluble material was removed by centrifugation at 2,600 × g for 20 min, and the extracted lipids were transferred with the supernatant to new tubes. The lipids were extracted again as described above; chloroform and buffer were added to combined extracts to obtain a methanol/chloroform/buffer ratio of 1:1:0.9. Following vigorous vortexing, samples were centrifuged at 2,600 × g for 20 min, and the chloroform phase containing lipids was transferred to a new tube. The lipids were dried under a stream of nitrogen, and the dry weight was determined. The lipids were then suspended in methanol-chloroform (1:1) at a concentration of 25 to 50 mg/ml and stored at −20°C. Membrane lipids from E. coli expressing YpfP (ANG373), from E. coli expressing YpfP and LtaA (ANG374), or from a control strain (ANG243) were extracted from 1-liter cultures. One liter of Luria-Bertani medium containing 100 μg/ml ampicillin was inoculated with 20 ml of an overnight culture, and the resulting culture was grown for 5 h at 37°C. Bacteria were collected by centrifugation and washed with 50 ml of 0.1 M sodium citrate (pH 4.7), and bacterial pellets were suspended in 8 ml of the same buffer. Bacteria were lysed by sonicating the preparations four times for 30 s at setting 7 using a Branson Sonifier 185 cell disruptor equipped with a microtip. Bacterial debris was collected by centrifugation at 100,000 × g for 30 min, and lipids were extracted as described above. Dried E. coli lipids were suspended in 1:1 in methanol-chloroform (1:1) at a concentration of 100 mg/ml, and 10-μl samples (1 mg lipid) were analyzed by thin-layer chromatography (TLC) and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry.
Lipid analysis by TLC.
Lipids were separated by TLC using Whatman Silica Gel A60 plates. Prior to separation, TLC plates were dried for 2 h at 100°C. Samples (300 to 1,000 μg lipid in 10 to 20 μl) were spotted on plates and separated using chloroform-methanol-H2O (70:30:4 or 65:25:4), as previously described (25-27). For detection of glycolipids, TLC plates were sprayed with 0.5% α-naphthol in 50% methanol and then with 95% H2SO4. The plates were subsequently incubated for 15 to 20 min at 100 to 120°C, which revealed glycolipids as pink spots (25-27). Digalactosyl-diacylglycerol (Sigma) was used as a chromatographic standard in some experiments. For structural analysis by MALDI-TOF mass spectrometry, glycolipids were purified following TLC by scraping silica gel from plates and extracting lipids twice with 3 ml of chloroform-methanol (1:1). Appropriate areas for lipid extraction were determined by developing one lane run in parallel with α-naphthol and H2SO4. Silica gel was sedimented by centrifugation at 2,600 × g for 10 min, and lipids in the methanol-chloroform extracts were transferred to new tubes. Lipids were recovered from the lower chloroform phase after the addition of H2O to obtain a chloroform-methanol-H2O (1:1:0.9) mixture, vortexing, and centrifugation for 10 min at 2,600 × g. Lipids were dried under a stream of nitrogen and prepared for MALDI-TOF analysis as described below. LTA glycolipid anchor structures were analyzed by TLC and were visualized with α-naphthol as described above. For nonglycolipid LTA anchor structures, lipids were separated on Silica Gel A60 plates using a heptane-isopropyl ether-acetic acid (60:40:4) solvent system; lipids were visualized by staining with a 0.2% amido black solution in 1 M NaCl, as previously described (40). 1,2-Dipalmitoyl-sn-glycerol (Avanti Polar Lipids, Inc.) was used for TLC calibration.
Lipid analysis by the MALDI-TOF method.
Dried lipids were suspended in 10 μl of a 0.125 M 2,2′:5′,2"-terthiophene (Sigma Aldrich) MALDI matrix dissolved in chloroform-methanol (1:1). Tenfold dilutions of the samples were prepared using the same MALDI matrix, and, where indicated below, NaHCO3 was added to a final concentration of 5 mM. Two 0.3-μl samples were spotted, and MALDI-TOF spectra were obtained with a Reflection time of flight instrument (ABI Biosystems) in the positive reflection mode.
LTA purification and lipid anchor isolation.
LTA from S. aureus was purified by hydrophobic interaction chromatography using a 5-ml HiTrap Octyl FF column (Amersham Biosciences). Four liters of TSB was inoculated with 40 ml of a wild-type or mutant S. aureus culture, and cultures were grown at 37°C to an OD600 of ∼3.5. Cultures were chilled on ice for 45 min, and bacteria were sedimented by centrifugation at 6,000 × g for 10 min and washed with 360 ml of 0.1 M sodium citrate (pH 4.7). Then the bacteria were sedimented by centrifugation at 8,000 ×g for 10 min, suspended in 40 ml of 0.1 M sodium citrate (pH 4.7), and disrupted with a bead beater (Biospec Products, Inc.) by shearing them five times for 2 min at 4°C with 0.1-mm glass beads (40 ml). The glass beads were sedimented by centrifugation for 1 min at 200 × g. Cell debris was collected by centrifugation and washed with 0.1 M sodium citrate (pH 4.7), and samples were stored at −20°C. Pellets were suspended in 0.1 M sodium citrate (pH 4.7) (0.4 g/ml) and stirred at room temperature for 30 min with an equal volume of 1-butanol. Insoluble material was sedimented by centrifugation at 13,000 × g for 20 min, extracts were transferred to new tubes, and phases were separated by centrifugation at 13,000 × g for 20 min. The aqueous (lower) phase containing LTA was retrieved. Pellets were reextracted with 1-butanol as described above, and LTA-containing phases were combined and extracted again with 1-butanol. Following phase separation, the volume of the LTA-containing phase was reduced to 6 to 7 ml using a Speed Vac, and the preparation was dialyzed against three changes of 1 liter of 20 mM sodium citrate (pH 4.7) using Spectra/Por 6 dialysis membranes (1,000-Da cutoff; Spectrum Laboratories, Inc.). Following dialysis, samples were manipulated so that each 10-ml sample contained 15% 1-propanol in 0.1 M sodium citrate (pH 4.7). Samples (3 ml) were loaded onto 5-ml HiTrap Octyl FF columns equilibrated with 0.1 M sodium citrate (pH 4.7)-15% 1-propanol. The columns were washed with 90 ml of 50 mM sodium citrate (pH 4.7)-15% 1-propanol buffer, and LTA was eluted with a linear 15 to 65% 1-propanol gradient in 50 mM sodium citrate (pH 4.7) in 90 ml. The flow rate was set at 1.5 ml/min, and 2-ml fractions were collected. Fractions from two runs were combined, aliquots were analyzed by immunoblotting for the presence of LTA, and positive fractions were pooled. For lipid anchor analysis, purified LTA was dialyzed extensively against H2O at 4°C and freeze-dried, and the dry weight was determined. Forty milligrams of LTA was hydrolyzed in 1 ml of 48% hydrofluoric acid (HF) for 44 to 48 h at 4°C. The reaction mixtures were neutralized with saturated NaHCO3, and lipids were extracted for 1 h at room temperature by addition of chloroform-methanol to obtain a final chloroform/methanol/aqueous phase ratio of 1:1:0.9. Samples were centrifuged for 10 min at 2,600 × g, and the chloroform phase, containing the LTA lipid anchor, was transferred to a new tube. The lipids were dried under a stream of nitrogen and suspended in a small volume of chloroform-methanol (1:1), and the lipid anchor from 10 mg LTA was analyzed by TLC or MALDI-TOF mass spectrometry.
Bacterial growth curves and murine infection and abscess formation.
For growth curves, three independent cultures of S. aureus strains Newman and ANG460 (ltaA mutant) were diluted 100-fold into 30 ml TSB and incubated at 37°C with shaking. At intervals culture aliquots were removed, and OD600 values were determined. The maximal doubling times during exponential growth (1.25 h to 3.75 h after inoculation) were calculated for wild-type and mutant strains. For virulence studies, strains Newman and ANG460 (ltaA mutant) were grown overnight in TSB at 37°C, diluted 100-fold into 25 ml fresh medium, and incubated at 37°C for 3 h. Staphylococci were collected by centrifugation, washed, and suspended in sterile phosphate-buffered saline. Bacterial suspensions were diluted to obtain an OD600 of 0.4, and 100-μl portions of the bacterial suspensions were administered intravenously via retroorbital injection to 45-day-old BALB/c mice (10 to 12 mice per group). Four days after injection, the mice were killed by CO2 asphyxiation, and the kidneys and livers were removed. The organs were homogenized in 1 ml of phosphate-buffered saline-1% Triton X-100, and dilutions of the homogenates were plated on tryptic soy agar plates to determine the number of CFU/ml. The detection limit was 33 CFU/ml, and the value for organs of animals with levels below the detection limit was defined as 32 CFU/ml for statistical analysis using a one-tailed Student's t test.
RESULTS
Genes required for glycolipid synthesis in S. aureus.
Membranes of S. aureus contain approximately 9% glycolipid (28); the predominant glycolipid, Glc2-DAG, is also used as a membrane anchor for LTA (8). Previous studies have shown that ypfP, which encodes a processive glycosyltransferase, is absolutely required for Glc2-DAG synthesis in S. aureus (27). YpfP employs UDP-glucose as a substrate for glycolipid synthesis (24, 27). In B. subtilis, two enzymes, encoded by pgcA and gtaB, are involved in UDP-glucose synthesis, using a pathway shown in Fig. 2A. BLAST searches with B. subtilis PgcA and GtaB as queries identified the S. aureus Mu50 genome-encoded SAV2491 (E-value, 2e-97) and SAV2500 (E-value, e-110) products as PgcA and GtaB homologs, respectively (Fig. 2A). Transposon insertions in the genes were isolated in a nonredundant collection of bursa aurealis mutants (2). Transposon insertions from original library strains ΦΝΞ177-2 (pgcA), ΦΝΞ1952 (gtaB), and ΦΝΞ171-39 (ypfP) were transduced into the protein A-deficient RN4220-derived laboratory strain SEJ1. Transposon insertion sites in the transductants, strains ANG372 (gtaB, SAV2941), ANG371 (pgcA, SAV2941), and ANG370 (ypfP, SAV1017), were confirmed by inverse PCR and DNA sequencing. To determine the requirement for S. aureus PgcA and GtaB for glycolipid synthesis, total membrane lipids were extracted from mutant and wild-type strains and analyzed by TLC, followed by α-naphthol/sulfuric acid staining of glycolipids. As expected from previous work (27, 28), ANG361 (wild-type parent), which has a transposon insertion in a gene irrelevant for glycolipid production, contained one prominent α-naphthol-reactive species that comigrated with the calibration standard, Gal2-DAG (Fig. 2B). The results of MALDI-TOF analysis of this compound were in agreement with predicted masses for Glc2-DAG (see below). Furthermore, Glc2-DAG was not detected in membrane extracts from ypfP mutant staphylococci. TLC analysis also revealed that Glc2-DAG was not present in membrane extracts obtained from pgcA or gtaB mutant staphylococci (Fig. 2B). In ypfP mutants, LTA with a wild-type chain length is anchored in the membrane via DAG (27). This structural change resulted in altered (slower) mobility of LTA on SDS-PAGE, as visualized by immunoblotting with polyglycerolphosphate-specific antibody (20). Similar changes in LTA motility were observed for LTA isolated from pgcA and gtaB mutant S. aureus strains (Fig. 2C). Together, these results show that pgcA and gtaB are essential for glycolipid synthesis in S. aureus and that in the absence of these genes LTA with an altered structure is produced. The mobility of LTA extracted from pgcA and gtaB mutant strains is identical to the mobility of LTA isolated from a ypfP mutant, suggesting that all of these LTAs have similar lipid anchor structures.
Mutations in ltaA cause structural changes in staphylococcal LTA.
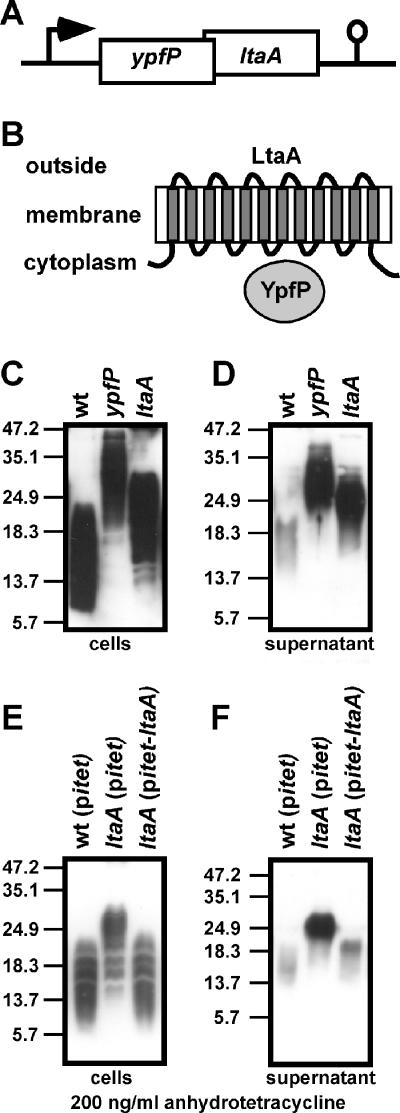
In S. aureus strains, ypfP is located in an operon with another gene, annotated in the MU50 genome by locus tag SAV1016 (designated ltaA here), whose predicted start site overlaps the 3′ end of the ypfP coding sequence (Fig. 3A). LtaA homologs (E-value, >5e-158) are found in all S. aureus strains sequenced and in closely related species (Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus saprophyticus) and are always preceded by ypfP. While YpfP is a cytoplasmic protein, LtaA is predicted to encode a 12-transmembrane-domain protein with an unknown function (Fig. 3B). LtaA encoded by the MU50 genome (Uniprot ID Q99V76) is annotated as a multidrug resistance-related protein, and multiple-sequence alignment using Pfam identified an MFS-1 domain in LtaA, which otherwise is found in members of the major facilitator superfamily clan (10). Genes that are located in an operon often encode proteins that function in the same biochemical pathway. To investigate a possible function of ltaA in LTA or glycolipid biosynthesis, we transduced a mutant allele (transposon insertion in ltaA) with phage Φ85 into strain SEJ1 to generate S. aureus strain ANG359. Cell-associated LTA extracted from S. aureus ANG359 migrated more slowly on an SDS-PAGE gel than LTA isolated from the wild-type strain but faster than LTA from a ypfP mutant (Fig. 3C). Similar to the observation for a ypfP mutant, an increase in the amount of LTA released into the culture supernatant was observed for the ltaA mutant (Fig. 3D) (27). To examine a possible polar effect of the transposon insertion on ypfP expression in strain ANG359, we performed a complementation analysis. DNA including ltaA was cloned into the single-site integration vector pitet containing an inducible tetracycline promoter (Pitet) inserted into pCL55. The resulting plasmid, pitet-ltaA, was integrated into the geh lipase gene of strain ANG359 to generate S. aureus ANG391. When this strain was grown in the presence of the Pitet inducer anhydrotetracycline, cell-associated LTA from ANG391 migrated with the same mobility on an SDS-PAGE gel as wild-type LTA (Fig. 3E). Furthermore, similar to the observation for a wild-type strain, only small amounts of LTA were released into the culture supernatant by the complemented strain (Fig. 3F). Thus, inactivation of ltaA causes structural changes in LTA that can be detected by SDS-PAGE, which prompted the designation ltaA (for lipoteichoic acid gene A).
FIG. 3.
(A) S. aureus ypfP operon. ypfP is located in an operon with ltaA (locus tag, SAV1016), which encodes a 12-transmembrane-segment protein predicted to function as a permease of the major facilitator superfamily. The diagram also shows a predicted transcriptional terminator downstream of ltaA. (B) Predicted subcellular locations of YpfP and LtaA. YpfP is a cytoplasmic protein, whereas LtaA is thought to reside in the cytoplasmic membrane. (C) Cell-associated LTA from S. aureus strains ANG361 (wild-type) (wt), ANG370 (ypfP), and ANG359 (ltaA) was extracted and normalized based on culture OD600 values, and the preparations were analyzed by immunoblotting. (D) Immunoblot analysis of LTA shed into the culture supernatant using the strains used for panel C. (E and F) For complementation analysis, a single-site chromosomal integration vector carrying ltaA under control of the tetracycline-inducible Pitet promoter was used (pitet-ltaA). S. aureus strains ANG395 (wild-type with empty vector pitet) (wt), ANG393 (ltaA mutant with empty vector pitet), and ANG391 (ltaA mutant with integrated pitet-ltaA) were grown in the presence of 200 ng/ml anhydrotetracycline. Cell-associated LTA (E) and LTA shed into the culture supernatant (F) were analyzed by immunoblotting. Samples were normalized based on culture OD600 values. The positions of protein molecular mass markers (in kDa) are indicated on the left.
Staphylococcal ltaA is not required for glycolipid synthesis.
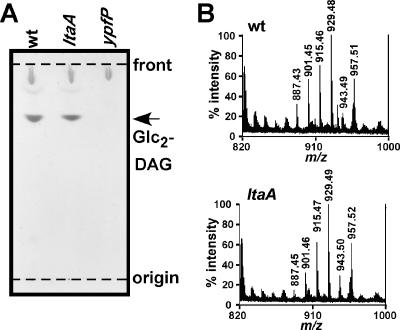
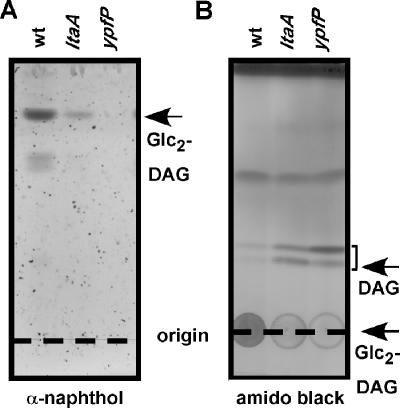
Glycolipids are not detectable in the membranes of ypfP, pgcA, or gtaB mutant S. aureus strains (see above). To investigate whether glycolipid production is affected in an ltaA mutant strain, total membrane lipids were extracted and separated by TLC, and glycolipids were visualized by α-naphthol/sulfuric acid staining. A major α-naphthol-reactive species was detected in membranes of ltaA mutant staphylococci, and this molecule migrated with the same mobility on TLC as Glc2-DAG from membranes of wild-type staphylococci (Fig. 4A). To verify the identity of the α-naphthol-reactive species, lipids were extracted from TLC plates and analyzed by MALDI-TOF mass spectrometry, and spectra were recorded in the reflector positive ion mode. The mass-to-charge (m/z) ratios of all major ion signals of the α-naphthol-reactive species were in agreement with the predicted mass of Glc2-DAG sodium adducts harboring fatty acids with chain lengths ranging from C15 to C18 (Fig. 4B) (i.e., observed m/z 929.5 and predicted m/z 929.6 for the sodium ion of Glc2-DAG harboring C18 and C15 acyl chains). The differences of 14 mass units in the spectra in Fig. 4 correspond to addition or omission of methylene (CH2) groups in Glc2-DAG. A complete list of observed and predicted compound masses, chemical structures, and possible acyl chain lengths for glycolipids in wild-type and ltaA mutant strains is provided in Table 2. When analyzing membranes of ypfP mutant staphylococci, we could not detect ion signals in the range from m/z 880 to m/z 960 (Table 2). In summary, our results reveal that inactivation of LtaA does not affect glycolipid synthesis in S. aureus.
FIG. 4.
(A) TLC analysis of S. aureus glycolipids. Membrane lipids were extracted from S. aureus strains ANG361 (wild-type) (wt), ANG359 (ltaA), and ANG370 (ypfP) and separated on TLC plates. Glycolipids were visualized with α-naphthol/sulfuric acid. The positions of the origin and solvent front are indicated by dashed lines. (B) MALDI-TOF mass spectrometry of S. aureus glycolipids. Glycolipids were isolated by TLC, extracted, dried, and suspended in 0.125 M 2,2′:5′,2"-terthiophene dissolved in chloroform-methanol (1:1). Spotted samples were dried and ionized by MALDI, and mass spectra were collected with a Reflection time of flight instrument in the positive ion mode.
TABLE 2.
Masses of diglucosyl-diacylglycerol lipids isolated from membranes of wild-type and mutant S. aureus strains
| Possible fatty acid chain length | Chemical formula | Predicted molecular mass | Observed molecular mass for:
|
||
|---|---|---|---|---|---|
| Wild type | ltaA mutant | ypfP mutant | |||
| C15/C15 | C45H84NaO15 | 887.57 | 887.43 | 887.45 | Absent |
| C15/C16 | C46H86NaO15 | 901.59 | 901.45 | 901.46 | Absent |
| C15/C17 | C47H88NaO15 | 915.60 | 915.46 | 915.47 | Absent |
| C15/C18 | C48H90NaO15 | 929.62 | 929.48 | 929.49 | Absent |
| C16/C18 | C49H92NaO15 | 943.63 | 943.49 | 943.50 | Absent |
| C17/C18 | C50H94NaO15 | 957.65 | 957.51 | 957.52 | Absent |
LtaA modulates YpfP-dependent glycolipid biosynthesis.
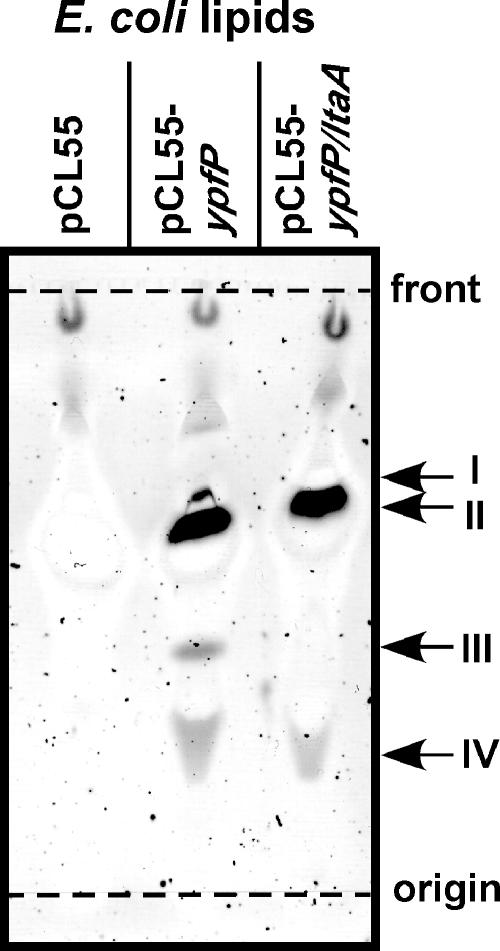
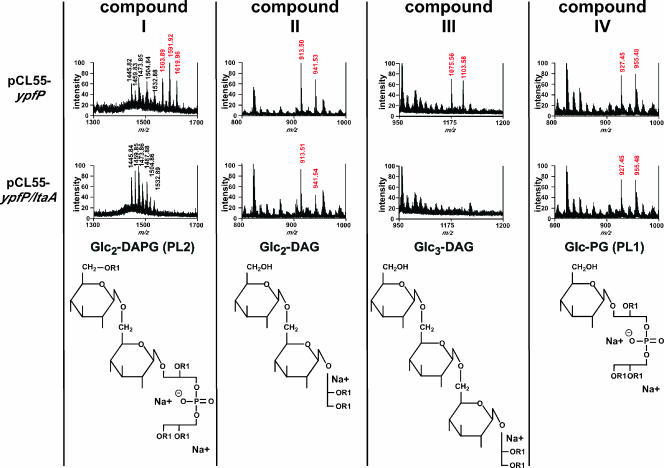
Previous work showed that expression of B. subtilis ypfP in E. coli leads to the production of glycolipids (25). Moreover, expression of S. aureus ypfP in E. coli not only generates Glc2-DAG but also leads to the formation of three other glycolipids, monoglycosyl-phosphatidylglycerol (Glc-PG) (also designated PL1), diglucosyl-diacyl-phosphatidylglycerol (Glc2-DAPG) (also designated PL2), and triglucosyl-diacylglycerol (Glc3-DAG) (24). In agreement with previous reports, we observed that expression of ypfP in E. coli ANG373(pCL55-ypfP) led to the production of four glycolipid species that could be detected by α-naphthol staining of TLC plates (compounds I to IV) (Fig. 5). As a control, glycolipids were not detected in membrane extracts of E. coli ANG243 harboring the empty vector pCL55 (Fig. 5). E. coli strain ANG374(pCL55-ypfP/ltaA) expressed both ypfP and ltaA. Only two glycolipid species, compounds II and IV, were detected in membranes of E. coli ANG374 (Fig. 5). To identify these compounds and to characterize the functions of LtaA in modulating YpfP-dependent glycolipid biosynthesis, lipids were purified from TLC plates and analyzed by MALDI-TOF mass spectrometry, and spectra were recorded in the positive reflector ion mode. The mass spectra collected for the four glycolipids (Glc2-DAG, Glc-PG, Glc2-DAPG, and Glc3-DAG) produced by E. coli strain ANG373(pCL55-ypfP) were in agreement with previously reported masses and proposed structures (24) (Fig. 6). Membranes of E. coli ANG374(pCL55-ypfP/ltaA), expressing both ltaA and ypfP, harbored only Glc2-DAG (compound II) and Glc-PG (compound IV) and not Glc2-DAPG (compound I) or Glc3-DAG (compound III) (Fig. 6). A complete list of the observed and predicted masses and chemical structures of glycolipids is provided in Table 3 and Fig. 6. Taken together, these experiments demonstrated that LtaA restricts the spectrum of YpfP-derived glycolipids to two compounds, Glc2-DAG and Glc-PG, and abrogates the formation of the aberrant products Glc2-DAPG and Glc3-DAG, which are also not found in membranes of S. aureus.
FIG. 5.
TLC analysis of glycolipids generated by YpfP in E. coli. Membrane lipids isolated from E. coli strains ANG243 (vector control; pCL55), ANG373 (plasmid expressing YpfP; pCL55-ypfP), and ANG374 (plasmid expressing YpfP and LtaA; pCL55-ypfP/ltaA) were separated by TLC, and glycolipids were detected by α-naphthol/sulfuric acid staining. Glycolipid species were designated compounds I to IV.
FIG. 6.
MALDI-TOF mass analysis of YpfP-generated glycolipids in E. coli. Lipids from E. coli were separated by TLC, and compounds I to IV were isolated. Dried lipids were suspended in 0.125 M 2,2,5′,2"-terthiophene-5 mM NaHCO3 and analyzed by using MALDI-TOF mass spectrometry, and mass spectra were collected in the positive ion mode. Mass-to-charge (m/z) ratios derived from glycolipid ion signals are indicated by red, and the corresponding structures (24) are shown at the bottom.
TABLE 3.
Masses of glucose-containing lipids isolated from E. coli membranes expressing YpfP or YpfP/LtaA
| Compound | Glycolipida | Chemical formula | Possible fatty acid chain length | Predicted molecular mass | Observed molecular mass for:
|
|
|---|---|---|---|---|---|---|
| YpfP | YpfP and LtaA | |||||
| I | PL2 (Glc2-DAPG)a | C82H150Na2O22P1 | 2×C16/2×C16:1 | 1,564.02 | 1,563.89 | Absent |
| C84H154Na2O22P | C16/2×C16:1/C18 | 1,592.05 | 1,591.92 | Absent | ||
| C86H158Na2O22P | C16/C16:1/C18/C18:1 | 1,620.08 | 1,619.96 | Absent | ||
| II | Glc2-DAG | C47H86NaO15 | C16/C16:1 | 913.59 | 913.50 | 913.51 |
| C49H90NaO15 | C16/C18:1 | 941.61 | 941.53 | 941.54 | ||
| III | Glc3-DAG | C53H96NaO20 | C16/C16:1 | 1,075.64 | 1,075.56 | Absent |
| C55H100NaO20 | C16/C18:1 | 1,103.67 | 1,103.58 | Absent | ||
| IV | PL1 (Glc-PG)a | C44H82Na2O15P | C16/C16:1 | 927.52 | 927.45 | 927.45 |
| C46H86Na2O15P | C16/C18:1 | 955.55 | 955.48 | 955.48 | ||
See reference 24.
The molecular details of YpfP-dependent biosynthesis of Glc2-DAPG (PL2) are currently not understood. Presumably, YpfP adds two glucose molecules to phosphatidylglycerol, whereas an E. coli acyltransferase may subsequently transfer additional fatty acids to generate Glc2-DAPG (24). Glc3-DAG is likely synthesized by YpfP via successive addition of thee glucose molecules to DAG, presumably without the aid of E. coli enzymes. Addition of the third glucose residue appears to be the rate-limiting step, as the amount of Glc2-DAG that accumulates in E. coli membranes is greater than the amount of Glc3-DAG that accumulates. We presume that LtaA prevents formation of Glc3-DAG by binding or sequestration of Glc2-DAG from YpfP. For example, LtaA may promote Glc2-DAG transport from the inner leaflet of the plasma membrane to the outer leaflet. If this is so, Glc2-DAG may not be accessible for YpfP, which is located in the cytoplasm. The transmembrane topology and predicted permease function of LtaA are certainly in agreement with this model.
Diacylglycerol anchor structures of LTA in ltaA mutant staphylococci.
Glycolipid synthesis occurs on membranes in the bacterial cytoplasm, whereas LTA biosynthesis is thought to be completed on membrane surfaces outside the cytoplasm (12). Thus, even though Glc2-DAG synthesis is not abolished in ltaA mutant staphylococci, mutations in ltaA could still affect the incorporation of Glc2-DAG as an LTA anchor. One could assume that LtaA functions as a permease for glycolipid transport across the cytoplasmic membrane. To test this hypothesis, we purified LTA molecules extracted from membranes of wild-type (ANG361), ypfP (ANG370), and ltaA (ANG359) staphylococcal strains and analyzed their membrane anchor structures. Purified LTA was treated with 48% HF to hydrolyze the phosphodiester bonds of the glycerolphosphate subunits. LTA membrane anchor lipids, which resist HF treatment, were subsequently extracted with chloroform-methanol and analyzed by TLC. As previously reported, Glc2-DAG is the predominant membrane anchor lipid of LTA in wild-type S. aureus strains (Fig. 7A). Small amounts of a lipid compound that comigrated with DAG were observed in HF-treated LTA samples from wild-type staphylococci (Fig. 7B). α-Naphthol-reactive glycolipids were not detected following HF treatment of LTA that had been purified from membranes of ypfP mutant staphylococci. Moreover, LTA anchor structures in ypfP mutant staphylococci were comprised of DAG. These findings are consistent with a model indicating that DAG or phosphatidylglycerol moieties retain biosynthetic LTA intermediates (polyglycerolphosphate polymerized via successive addition of glycerolphosphate derived from phosphatidylglycerol) in the membrane until transfer to Glc2-DAG completes the synthesis pathway (5). However alternative models for LTA synthesis have been proposed, in which glycerolphosphate is directly polymerized on glycolipids (11, 19) and DAG-linked polyglycerolphosphate LTA may therefore be synthesized only in the absence of glycolipids. HF treatment of LTA purified from membranes of ltaA mutant strains released small amounts of glycolipids and also additional anchor lipids (Fig. 7). Compared to the LTA from the wild-type parent, the LTA from the ltaA mutant contained decreased amounts of Glc2-DAG and increased amounts of DAG (Fig. 7). A mixture of glycolipid and nonglycolipid anchor structures is in agreement with our initial observation that there were discrete differences in mobility on SDS-PAGE gels between LTA isolated from the S. aureus ltaA mutant strain and LTA isolated from wild-type (Glc2-DAG anchor) and ypfP mutant (DAG anchor) strains. Together, these results indicate that even though LtaA is not required for glycolipid synthesis, the predicted membrane permease is required for LTA attachment to Glc2-DAG anchor structures.
FIG. 7.
LTA membrane anchor structures. LTA from S. aureus strains ANG361 (wild-type) (wt), ANG359 (ltaA), and ANG370 (ypfP) was purified, and polyglycerolphosphate was hydrolyzed with hydrofluoric acid. LTA anchor structures, which resist hydrofluoric acid treatment, were isolated by organic solvent extraction and analyzed by TLC. (A) LTA anchor structures were separated by TLC using chloroform-methanol-H2O (70:30:4), and glycolipids visualized with α-naphthol/sulfuric acid. (B) Lipids were separated using heptane-isopropyl ether-acetic acid (60:40:4) and were visualized by staining with a 0.2% amido black solution. The chromatographic mobilities of diacylglycerol (1,2-dipalmitoyl-sn-glycerol) and Glc2-DAG are indicated on the right.
S. aureus mutants lacking ltaA display a virulence defect in a murine abscess model.
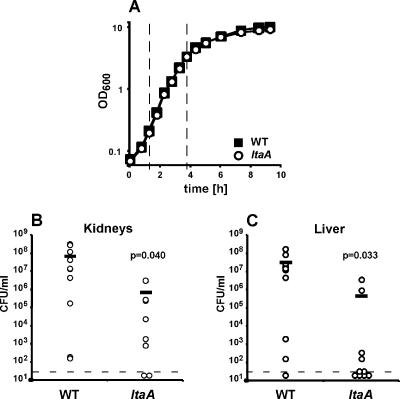
To assess whether glycolipid anchoring of LTA is required for the pathogenesis of staphylococcal infections, an ltaA mutant was constructed using the S. aureus human clinical isolate Newman (9). LTA isolated from the Newman-derived ltaA mutant ANG460 displayed an electrophoretic mobility shift similar to the shift displayed by LTA isolated from the RN4220-derived mutant, indicating that strain ANG460 is also defective in glycolipid anchoring of LTA (data not shown). Wild-type strain Newman and the isogenic ltaA mutant replicated with similar doubling times (38 and 37 min, respectively) during logarithmic growth in TSB (Fig. 8A). The ltaA mutant had a slightly lower density during the postexponential growth phase (Fig. 8A), which may have coincided with changes in membrane lipid composition that are known to occur during later growth stages (28, 45). For virulence studies, 45-day-old BALB/c mice were infected with S. aureus strain Newman or an isogenic variant lacking ltaA. Infected animals were killed after 4 days, the livers and kidneys were removed, and homogenized tissues were plated on agar plates to determine staphylococcal replication in organ abscesses. Compared to the replication of the wild-type parent, the replication of the staphylococcal variant lacking ltaA was significantly impaired in kidney and liver tissues (Fig. 8B and C). On balance, we observed a 2-log reduction in the number of CFU recovered, suggesting that glycolipid anchoring of LTA plays an important role in the functional assembly of staphylococcal cell wall envelopes during infection.
FIG. 8.
S. aureus mutants lacking ltaA display a virulence defect in the murine abscess model. (A) Growth curves for S. aureus strain Newman and an isogenic ltaA mutant. Overnight cultures of S. aureus wild-type strain Newman (WT) and ltaA mutant strain ANG460 were diluted into fresh TSB, and the cultures were incubated at 37°C. At intervals samples were withdrawn, and OD600 values were determined. Average values for three independently grown cultures are shown. During logarithmic growth (1.25 to 3.75 h after inoculation, indicated by dashed lines) the wild-type and ltaA mutant strains had doubling times of 38 and 37 min, respectively. (B and C) Virulence test with S. aureus strain Newman and an isogenic ltaA mutant. BALB/c mice were challenged by retroorbital infection with S. aureus strain Newman or ANG460 (ltaA). Four days after infection, the animals were killed, and the kidneys and livers were removed. Infected organ tissues were homogenized and spread on agar plates, and the staphylococcal burden was quantified by using colony formation and enumeration. The dashed lines indicate the limits of detection (33 CFU/ml), and the horizontal bars indicate observation averages. The statistical significance of the reduction in the staphylococcal burden was assessed with the one-tailed Student t test, and P values were determined.
DISCUSSION
Despite rigorous biochemical analysis of LTA, the genetic determinants required for LTA synthesis have still not been defined. The dlt operon, encoding factors that catalyze d-alanyl esterification of glycerolphosphate and ribitolphosphate teichoic acids, has been characterized for several different gram-positive bacteria (22, 36, 37). d-Alanyl esterification of teichoic acids plays an important role during host-pathogen interactions of S. aureus, group A streptococci, Listeria monocytogenes, and B. anthracis (1, 17, 30, 39). Only one additional gene has been reported to be required for LTA biosynthesis in S. aureus. ypfP, which encodes a processive glycosyltransferase, is required for glycolipid and LTA anchor structure synthesis (27). Here we describe identification of three additional S. aureus genes, pgcA, gtaB, and ltaA, which are involved in the synthesis of LTA.
B. subtilis pgcA and gtaB encode enzymes required for the synthesis of UDP-glucose, which is used by YpfP as a substrate for glycolipid synthesis (32, 41, 46). Using bioinformatic analysis, we identified S. aureus PgcA and GtaB homologs, and here we present evidence that these proteins are also required for glycolipid biosynthesis in S. aureus (Fig. 2). In B. subtilis, UDP-glucose has a central role as a metabolite for the synthesis of cell wall envelope polymers. UDP-glucose is required not only for glycolipid synthesis but also for glycosylation of the major wall teichoic acid and for the synthesis of the minor wall teichoic acid (32). In addition, UDP-glucose plays a central role in B. subtilis biofilm formation through a pathway that is distinct from the requirement for UDP-glucose for glycolipid or wall teichoic acid synthesis (3, 32). Additional work is needed to investigate the requirement for pgcA and gtaB in S. aureus cell wall envelope assembly, cell shape determination, or biofilm formation.
Previous studies have shown that pgcA, gtaB, and ypfP mutant Bacillus strains, as well as ypfP mutant S. aureus strains, have aberrant cell shapes (18, 27, 32, 42). It is not yet clear whether such morphological alterations are caused by a lack of membrane glycolipids or by changes in LTA structure or function (for example, by increased release of LTA into the culture media of ypfP mutant staphylococci) (27). Experimental distinction between these possibilities may now be possible if workers investigate gtaB, pgcA, ypfP, and ltaA mutant staphylococci. To our knowledge, the S. aureus ltaA mutant described in this report is the first mutant capable of segregating glycolipid and LTA biosynthesis. Membrane glycolipids are synthesized in an ltaA mutant strain in a fashion similar to that in wild-type staphylococci; however, only a small amount of LTA is anchored by Glc2-DAG, whereas a large portion is tethered to DAG, similar to LTA anchor structures in ypfP mutant membranes. These findings place LtaA activity downstream of the PgcA, GtaB, and YpfP activities in the LTA synthesis pathway outlined in Fig. 2A.
Glycolipid synthesis occurs in the bacterial cytoplasm, whereas LTA synthesis is thought to localize in the outer leaflet of the plasma membrane (12). We assume that spontaneous distribution of glycolipids from inner membrane leaflets to outer membrane leaflets occurs at a low rate. Transport of glycolipids by a permease may be required to achieve the necessary distribution and availability of glycolipids for LTA biosynthesis. ltaA codes for a hydrophobic protein with 12 predicted transmembrane domains. On the basis of Pfam amino acid sequence analysis, LtaA seems to be a member of the major facilitator superfamily clan (10). All members of the major facilitator superfamily possess either 12 or 14 transmembrane helices, and these proteins are membrane transporters that are ubiquitous in bacteria, archaea, and eukarya and function primarily in uniport, symport, or antiport of small solutes (38, 44). More recently, E. coli LplT, another member of the major facilitator superfamily, has been reported to catalyze the transmembrane movement of lysophospholipids from the membrane surface into the cytoplasm, where the lipid is acylated by a second protein encoded in the same operon (21). In the absence of LplT, lysophospholipids can still be incorporated into cells, albeit at a much lower rate, which presumably represents spontaneous flipping of lysophospholipid across membranes (21). These observations resemble our observations for LTA biosynthesis. In ltaA mutant strains, only small amounts of glycolipids may flip to the outside, as very little Glc2-DAG is found in LTA anchor structures (Fig. 7). If one also assumes that addition of Glc2-DAG to LTA continuously removes glycolipids from the outer leaflet of the plasma membrane, the slow diffusion of glycolipids across membranes may not fulfill the concentration requirements for glycolipid substrates in LTA biosynthesis. LtaA may therefore promote transfer of glycolipids across membranes following a concentration gradient.
A model in which LtaA functions as glycolipid flippase can also explain the role of LTA in modulating YpfP-dependent glycolipid synthesis in E. coli. In the absence of LtaA, but not in its presence, YpfP synthesizes Glc3-DAG in E. coli membranes. The addition of the third glucose molecule to Glc2-DAG is slow and rate limiting. If one assumes that LtaA mediates transport of Glc2-DAG across the membrane, this mechanism may prevent YpfP-mediated synthesis of Glc3-DAG in the bacterial cytoplasm. Nevertheless, our model cannot explain why YpfP-synthesized PL2 glycolipid with four acyl chains accumulates in the absence of LtaA but not in the presence of LtaA.
Even though in our study we could identify three S. aureus genes involved in glycolipid synthesis and LTA anchoring, other genes that are involved in LTA biosynthesis still remain to be discovered. Knowledge of the entire biosynthetic pathway is essential to increase our understanding of cell wall envelope assembly in gram-positive bacteria.
Acknowledgments
We thank Taeok Bae for construction of S. aureus strain SEJ1, Christiaan van Ooij, Lauriane Quenee, Anthony Maresso, Joe Sorg, Yukiko Stranger-Jones, and Luciano Marraffini for critical reading of the manuscript, and members of our laboratory for helpful discussions. We thankfully acknowledge Yukiko Stranger-Jones and Nancy Ciletti for their help with the animal experiment.
The study of lipoteichoic acid synthesis provided important insight into the assembly of surface proteins in the staphylococcal cell wall, work that was supported by Public Health Service grants AI38897 and AI52474 from the National Institute of Allergy and Infectious Diseases Division of Microbiology and Infectious Diseases to O.S. O.S. acknowledges membership in and support received from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (National Institute of Allergy and Infectious Diseases award 1-U54-AI-057153).
Footnotes
Published ahead of print on 5 January 2007.
REFERENCES
- 1.Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. [DOI] [PubMed] [Google Scholar]
- 2.Bae, T., A. K. Banger, A. Wallace, E. M. Glass, F. Aslund, O. Schneewind, and D. M. Missiakas. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA 101:12312-12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branda, S. S., J. E. Gonzalez-Pastor, E. Dervyn, S. D. Ehrlich, R. Losick, and R. Kolter. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 186:3970-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Button, D., and N. L. Hemmings. 1976. Teichoic acids and lipids associated with the membrane of a Bacillus licheniformis mutant and the membrane lipids of the parental strain. J. Bacteriol. 128:149-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu, T. H., H. Morimoto, and J. J. Baker. 1993. Biosynthesis and characterization of phosphatidylglycerophosphoglycerol, a possible intermediate in lipoteichoic acid biosynthesis in Streptococcus sanguis. Biochim. Biophys. Acta 1166:222-228. [DOI] [PubMed] [Google Scholar]
- 6.Cleveland, R. F., J. V. Höltje, A. J. Wicken, A. Tomasz, L. Daneo-Moore, and G. D. Shockman. 1975. Inhibition of bacterial wall lysins by lipoteichoic acids and related compounds. Biochem. Biophys. Res. Commun. 67:1128-1135. [DOI] [PubMed] [Google Scholar]
- 7.Doran, K. S., E. J. Engelson, A. Khosravi, H. C. Maisey, I. Fedtke, O. Equils, K. S. Michelsen, M. Arditi, A. Peschel, and V. Nizet. 2005. Blood-brain barrier invasion by group B Streptococcus depends upon proper cell-surface anchoring of lipoteichoic acid. J. Clin. Investig. 115:2499-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duckworth, M., A. R. Archibald, and J. Baddiley. 1975. Lipoteichoic acid and lipoteichoic acid carrier in Staphylococcus aureus H. FEBS Lett. 53:176-179. [DOI] [PubMed] [Google Scholar]
- 9.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 10.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, W. 1990. Bacterial phosphoglycolipids and lipoteichoic acids, p. 123-234. In D. Hanahan (ed.), Handbook of lipid research, vol. 6. Plenum Press, New York, NY. [Google Scholar]
- 12.Fischer, W. 1994. Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus. Med. Microbiol. Immunol. 183:61-76. [DOI] [PubMed] [Google Scholar]
- 13.Fischer, W. 1988. Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 29:233-302. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, W., T. Mannsfeld, and G. Hagen. 1990. On the basic structure of poly(glycerophosphate) lipoteichoic acids. Biochem. Cell Biol. 68:33-43. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, W., and P. Rosel. 1980. The alanine ester substitution of lipoteichoic acid (LTA) in Staphylococcus aureus. FEBS Lett. 119:224-226. [DOI] [PubMed] [Google Scholar]
- 16.Fischer, W., P. Rosel, and H. U. Koch. 1981. Effect of alanine ester substitution and other structural features of lipoteichoic acids on their inhibitory activity against autolysins of Staphylococcus aureus. J. Bacteriol. 146:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, N., L. Shetron-Rama, A. Herring-Palmer, B. Heffernan, N. Bergman, and P. Hanna. 2006. The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J. Bacteriol. 188:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsberg, C. W., P. B. Wyrick, J. B. Ward, and H. J. Rogers. 1973. Effect of phosphate limitation on the morphology and wall composition of Bacillus licheniformis and its phosphoglucomutase-deficient mutants. J. Bacteriol. 113:969-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganfield, M. C., and R. A. Pieringer. 1980. The biosynthesis of nascent membrane lipoteichoic acid of Streptococcus faecium (S. faecalis ATCC 9790) from phosphatidylkojibiosyl diacylglycerol and phosphatidylglycerol. J. Biol. Chem. 255:5164-5169. [PubMed] [Google Scholar]
- 20.Gründling, A., and O. Schneewind. 2006. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harvat, E. M., Y. M. Zhang, C. V. Tran, Z. Zhang, M. W. Frank, C. O. Rock, and M. H. Saier, Jr. 2005. Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J. Biol. Chem. 280:12028-12034. [DOI] [PubMed] [Google Scholar]
- 22.Heaton, M. P., and F. C. Neuhaus. 1992. Biosynthesis of d-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the d-alanine-activating enzyme. J. Bacteriol. 174:4707-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji, Y., B. Zhang, S. F. Van, Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 24.Jorasch, P., D. C. Warnecke, B. Lindner, U. Zahringer, and E. Heinz. 2000. Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids and glycosylsterols. Eur. J. Biochem. 267:3770-3783. [DOI] [PubMed] [Google Scholar]
- 25.Jorasch, P., F. P. Wolter, U. Zahringer, and E. Heinz. 1998. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol. Microbiol. 29:419-430. [DOI] [PubMed] [Google Scholar]
- 26.Kates, M. 1986. Techniques of lipidology, p. 347-446. In R. Burdon and P. van Knippenberg (ed.), Laboratory techniques in biochemistry and molecular biology, 2nd ed. American Elsevier, New York, NY.
- 27.Kiriukhin, M. Y., D. V. Debabov, D. L. Shinabarger, and F. C. Neuhaus. 2001. Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J. Bacteriol. 183:3506-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch, H. U., R. Haas, and W. Fischer. 1984. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur. J. Biochem. 138:357-363. [DOI] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 30.Kristian, S. A., V. Datta, C. Weidenmaier, R. Kansal, I. Fedtke, A. Peschel, R. L. Gallo, and V. Nizet. 2005. d-Alanylation of teichoic acids promotes group A streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J. Bacteriol. 187:6719-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert, P. A., I. C. Hancock, and J. Baddiley. 1977. Occurrence and function of membrane teichoic acids. Biochim. Biophys. Acta 472:1-12. [DOI] [PubMed] [Google Scholar]
- 32.Lazarevic, V., B. Soldo, N. Medico, H. Pooley, S. Bron, and D. Karamata. 2005. Bacillus subtilis α-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 71:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 34.Morath, S., A. Geyer, and T. Hartung. 2001. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193:393-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morath, S., S. von Aulock, and T. Hartung. 2005. Structure/function relationships of lipoteichoic acids. J. Endotoxin Res. 11:348-356. [DOI] [PubMed] [Google Scholar]
- 36.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuhaus, F. C., M. P. Heaton, D. V. Debabov, and Q. Zhang. 1996. The dlt operon in the biosynthesis of d-alanyl-lipoteichoic acid in Lactobacillus casei. Microb. Drug Resist. 2:77-84. [DOI] [PubMed] [Google Scholar]
- 38.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Götz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 40.Plekhanov, A. Y. 1999. Rapid staining of lipids on thin-layer chromatograms with amido black 10B and other water-soluble stains. Anal. Biochem. 271:186-187. [DOI] [PubMed] [Google Scholar]
- 41.Pooley, H. M., D. Paschoud, and D. Karamata. 1987. The gtaB marker in Bacillus subtilis 168 is associated with a deficiency in UDP-glucose pyrophosphorylase. J. Gen. Microbiol. 133:3481-3493. [DOI] [PubMed] [Google Scholar]
- 42.Price, K. D., S. Roels, and R. Losick. 1997. A Bacillus subtilis gene encoding a protein similar to nucleotide sugar transferases influences cell shape and viability. J. Bacteriol. 179:4959-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajbhandary, U. L., and J. Baddiley. 1963. The intracellular teichoic acid from Staphylococcus aureus H. Biochem. J. 87:429-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saier, M. H., Jr., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne, S. C. Huang, D. L. Jack, P. S. Jahn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T. T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 45.Short, S. A., and T. C. White. 1970. Metabolism of the glucosyl diglycerides and phosphatidylglucose of Staphylococcus aureus. J. Bacteriol. 104:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soldo, B., V. Lazarevic, P. Margot, and D. Karamata. 1993. Sequencing and analysis of the divergon comprising gtaB, the structural gene of UDP-glucose pyrophosphorylase of Bacillus subtilis 168. J. Gen. Microbiol. 139:3185-3195. [DOI] [PubMed] [Google Scholar]