Abstract
Faithful partitioning of genetic material during cell division requires accurate spatial and temporal positioning of nuclei within dividing cells. In Saccharomyces cerevisiae, nuclear positioning is regulated by an elegant interplay between components of the actin and microtubule cytoskeletons. Regulators of this process include Bud6p (also referred to as the actin-interacting protein Aip3p) and Kar9p, which function to promote contacts between cytoplasmic microtubule ends and actin-delimited cortical attachment points. Here, we present the previously undetected association of Bud6p with the cytoplasmic face of yeast spindle pole bodies, the functional equivalent of metazoan centrosomes. Cells lacking Bud6p show exaggerated movements of the nucleus between mother and daughter cells and display reduced amounts of time a given spindle pole body spends in close association with the neck region of budding cells. Furthermore, overexpression of BUD6 greatly enhances interactions between the spindle pole body and mother-bud neck in a spindle alignment-defective dynactin mutant. These results suggest that association of either spindle pole body with neck components, rather than simply entry of a spindle pole body into the daughter cell, provides a positive signal for the progression of mitosis. We propose that Bud6p, through its localization at both spindle pole bodies and at the mother-bud neck, supports this positive signal and provides a regulatory mechanism to prevent excessive oscillations of preanaphase nuclei, thus reducing the likelihood of mitotic delays and nuclear missegregation.
Regulation of spindle orientation and positioning is necessary for the accurate segregation of genomic material and plays an important role in asymmetric cell division (39). The effect of spindle rotation on cell cleavage is of particular importance during early stages of embryonic development when asymmetric cell divisions predict the fate of daughter cells (11, 13, 23). In Saccharomyces cerevisiae, cortical cues mark the position for bud growth in G1 and thereby determine the orientation of the spindle to be along the mother-bud axis and perpendicular to the cleavage plane. Considering the level of conservation of molecular factors involved in spindle positioning and the asymmetry of cell divisions, S. cerevisiae is an excellent model for studying this process (18, 26, 45).
In S. cerevisiae, nuclear migration and spindle alignment are governed by the complex interplay of overlapping pathways (26). In order for these pathways to culminate in an asymmetric orientation of the spindle and accurate chromosome segregation, the cell must regulate the activities of these pathways in a precise temporal and spatial program. Although some aspects remain hotly contested, the major elements have been recently well summarized (26) and are briefly reiterated here. In the earliest stages of G1, the cell has a single spindle pole body (SPBold) with nucleated astral microtubules. The actin-interacting protein Bud6p (also referred to as Aip3p) localizes to the incipient bud site at the earliest stages of G1 (5) and can be observed to capture and induce shortening of the astral microtubules emanating from the SPBold, resulting in orientation of SPBold toward the soon to be emerging bud (52). At the G1/S boundary, SPB duplication is completed coincident with bud emergence (60). Due possibly in part to the conservative nature of SPB duplication (9), SPB asymmetry is rapidly established via two known mechanisms. First, transient inhibition of new microtubule nucleation by the cyclin-dependent kinase Cdc28p/Clb5p is thought to restrict astral microtubules solely to SPBold (53); second, Kar9p localization is established and restricted to SPBold (35, 37). The astral microtubules nucleated solely off SPBold are captured at the bud cortex at sites of Bud6p localization (52), thereby reinforcing ultimate segregation of SPBold to the daughter cell. This polarity is further reinforced as the Kar9p-Bim1p complex (initially restricted to SPBold) is moved to the plus ends of microtubules, presumably by the action of the Kip2p kinesin (37). The Myo2p motor then captures Kar9p via its cargo-binding tail domain and pulls these microtubule plus ends toward the bud cortex along longitudinally oriented actin cables (7, 63). These actin cables result from actin filament nucleation at the bud cortex by the formin Bni1p (46, 48, 49) and its activator, Bud6p (42). The importance of the actin cytoskeleton for the early positioning of SPBold is underscored by the spindle-positioning defects seen in mutants lacking Bni1p, Bud6p, or other actin-regulatory factors (32, 41, 47, 59, 62).
As the bud enlarges, Bud6p localization shifts from the bud cortex to the neck (5), where it can again be seen to capture microtubules nucleated from SPBold (25, 51); these interactions are thought to help stabilize orientation of the spindle. Finally, dynein motor activity (along with Arp1p and the dynactin complex) utilizing the Num1p cortical anchor for purchase is believed to pull and elongate the spindle through the neck in preparation for anaphase (12, 19, 34, 44, 54, 58, 61). Additional contributions to spindle position are made by three kinesin-related proteins, Kip2p, Kip3p, and Kar3p (15-17, 40, 50).
As indicated above, Bud6p displays a cell-cycle-regulated localization pattern. At START, it localizes to the incipient bud site and remains solely at the bud cortex until G2/M. It then begins to accumulate in the bud neck, where it remains until initiation of the next cell cycle (5). The Cdc42p effector Gic2p (GTPase interactive component) functions in recruiting Bud6p and Bni1p to activated Cdc42p, thereby initiating actin polarization (29). Shortly after bud emergence, Gic2p is degraded by the ubiquitin-proteasome pathway (28), but Bud6p continues to be transported to polarity sites in association with secretory vesicles via the actin-based motor protein Myo2p (31).
We report here that Bud6p also localizes to the SPBs and that this localization is dependent on Cnm67p, the most central component of the SPB outer plaque. This new aspect of the Bud6p localization pattern led us to investigate whether potential interactions between SPBs and the bud neck contribute to preanaphase spindle dynamics. Time-lapse analysis of green fluorescent protein (GFP)-bud6p-1 (a noncomplementing C-terminal truncation mutant) behavior in wild-type cells allowed us to examine SPB movement relative to Bud6p structures at the bud neck. This analysis showed extended periods of association between either SPB and the neck region. Furthermore, SPB-neck interactions appear to define a barrier that prevents the spindle from being pulled completely through the neck. In the absence of functional Bud6p, this apparent barrier is lost and preanaphase nuclei can be observed to swing completely through the neck into the mother or daughter cell bodies. In addition, cells bearing a bud6Δ allele progress more slowly through the G2/M phase of the cell cycle than do wild-type cells, possibly due to decreased nucleus-neck interactions prior to anaphase. Overexpression of BUD6, on the other hand, can accentuate SPB-neck interactions. Collectively, our observations have led us to propose a model whereby Bud6p stabilizes SPB-neck interactions to hold the nucleus in position for accurate segregation through mitosis and that these interactions also provide positive signals for progression into anaphase.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The yeast strains used in this study are listed in Table 1. Strains originating from this study are in the S288C background. Standard methods were used for yeast growth, sporulation, and tetrad dissection (4). Yeast transformations were performed using the lithium acetate procedure (4).
TABLE 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| FY23x86 | MATa/MATα ura3-52/ura3-52 trp1Δ63/TRP1 HIS3/his3Δ200 leu2Δ1/leu2Δ1 | 31 |
| AHY1 | MATa/MATα ura3-52/ura3-52 trp1Δ63/TRP1 HIS3/his3Δ200 SPC42-ECFP::LEU2/leu2Δ1 | This study |
| AHY4 | MATα ura3-52 TRP1 his3Δ200 SPC42-ECFP::LEU2 | This study |
| AHY5 | MATaura3-52 trp1Δ63 his3Δ200 SPC42-ECFP:LEU2 | This study |
| AHY9 | MATaura3-52 trp1Δ63 HIS3 leu2Δ1 cnm67::kanR | This study |
| AHY13 | MATaura3-52 TRP1 his3Δ200 SPC42-ECFP:LEU2 cnm67::kanR | This study |
| AHY26 | MATa/MATα GFP-BUD6:URA3/ura3-52 trp1Δ63/TRP1 HIS3/his3Δ200 leu2Δ1/leu2Δ1 | This study |
| BHY443 | MATa/MATα ura3-52 his3Δ200 trp1Δ63 leu2Δ1 bud6-Δ2::HIS3 CDC14-GFP::kanR | This study |
| BHY444 | MATaura3-52 leu2Δ1 CDC14-GFP::kanr | This study |
| BHY482 | MATaura3Δ0 leu2Δ0 his3Δ1 bub2Δ::natRCDC14-GFP::kanr | This study |
| DAY101x102 | MATa/MATα ura3-52/ura3-52 trp1Δ63/trp1Δ63 his3Δ200/his3Δ200 bud6-Δ2::HIS3/bud6-Δ2::HIS3 leu2Δ1/leu2Δ1 | 5 |
| PY3643x3297 | MATa/MATα ura3Δ0/ura3Δ0 his3Δ1/his3Δ1 leu2Δ0/LEU2 LYS2/lys2Δ0 met15 Δ0/MET15 dyn1::kanR/dyn1::kanR | This study |
| YTC240 | MATalys2-801 his3-200 leu2-3,112 ura3-52 trp1-1 CDC14-GFP::kanRnme1-2::TRP1 pMES127[URA3 CEN NME1] | 10 |
| IAY522 | MATaho ura3 his3-11,15 trp1-leu2-3,112 ade2-1 1 can1-100 ssd1 nud1Δ::HIS5 NUD1::TRP1 | J. Kilmartin |
| IAY520 | MATaho ura3 his3-11,15 trp1-1 leu2-3,112 ade2-1 1 can1-100 ssd1 nud1Δ::HIS5 nud1-44::TRP1 | J. Kilmartin |
| YJC3677 | MATaLEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3681 | MATaarp1Δ::kanRLEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3685 | MATakar9Δ::hygbRLEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3689 | MATabud6Δ::His3MX6 LEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3693 | MATaarp1Δ::kanRkar9Δ::hygbRLEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3697 | MATaarp1Δ::kanRbud6Δ::His3MX6 LEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3701 | MATakar9Δ::hygbRbud6Δ::His3MX6 LEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC3705 | MATaarp1Δ::kanRkar9Δ::hygbRbud6Δ::His3MX6 LEU2::LEU2-GFP-TUB1 ura3-52 lys2-801 leu2-Δ1 his3-Δ200 | This study |
| YJC4035 | MATaLEU2::LEU2-GFP-TUB1 URA3::URA3-GAL1-BUD6-GST arp1Δ::kanRura3-52 lys2-801 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
hygb, hygromycin B gene.
AHY1 was constructed by integrating NruI-linearized pAH1 into the leu2 locus of the diploid strain FY23x86. The presence of the SPC42-ECFP (enhanced cyan fluorescent protein) cassette was confirmed by microscopic analysis. AHY4 and AHY5 were constructed by sporulation and tetrad dissection of AHY1. In all tetrads, the LEU2 marker cosegregated with SPC42-ECFP. The presence of the LEU2::SPC42-ECFP cassette did not affect growth compared to the wild type. AHY9 was constructed by double-fusion PCR (3) and integration of the resulting cnm67::Kanr cassette at the CNM67 locus of FY23x86. The first PCR amplified the CNM67 promoter region with primers AHo-CNM67-1 (5′-GACCATGAAAAATCTATGGTA-3′) and AHo-CNM67-2 (5′-TTAATTAACCCGGGGATCCGGATGTACAAAAGACCTGTCAC-3′) from genomic DNA. The second PCR amplified the kanamycin resistance (kanr) gene with primers F1 (5′-CGGATCCCCGGGTTAATTAA-3′) and R1 (5′-GAATTCGAGCTCGTTTAAAC-3′) using pFA6a-kanMX6 (36) as a template. The third PCR amplified the CNM67 terminator region with primers AHo-CNM67-3 (5′-GTTTAAACGAGCTCGAATTCATCCTGGAGAAGATGGTGAAG-3′) and AHo CNM67-4 (5′-GCTGGTCAGACTTTTACTATG-3′) from genomic DNA.
Transformants that had successfully integrated the cnm67::kanR cassette were isolated based on (i) their ability to grow in media supplemented with G418 (200 mg/liter; Invitrogen, Carlsbad, CA) and (ii) PCR analysis to verify heterozygosity for the CNM67 loci (CNM67/cnm67::kanR). Subsequent sporulation and dissection of positive candidates created AHY9. All tetrads segregated 2:2 for all markers tested; Kanr cosegregated with a slow-growth phenotype, in line with the previously reported growth behavior of a CNM67 null mutant (2). AHY13 was created by sporulation and tetrad dissection of AHY9x4.
Plasmid constructions and DNA manipulations.
Plasmids are listed in Table 2. Plasmid pDP128 (gift from David Pruyne) was constructed by cloning the GFP-BUD6-containing EcoRI-SalI fragment from pDAb204 into pRS306. For genome integration at the ura3-52 locus, pDP128 was digested with StuI and transformed into yeast selecting for Ura+ colonies (strain AHY26). Plasmid pAH1 was constructed by digesting pXH133 (gift from Peter Sorger) with PvuI and cloning the SPC42-ECFP insert into PvuI-digested pRS305. pAH1 was linearized with NcoI to promote integration at the leu2 locus upon transformation into yeast.
TABLE 2.
Plasmids used in this study
| Plasmid | Markers | Source or reference |
|---|---|---|
| pDP128 | Act1p-GFP-BUD61-788-Act1t URA3 | D. Pruyne |
| pXH133 | URA3 SPC42-ECFP | P. Sorger |
| pAH1 | LEU2 SPC42-ECFP | This study |
| pTD125 | Act1p-GFP-MCS-Act1t URA3 Cen | T. Doyle |
| pDAb204 (pRB2190) | Act1p-GFP-BUD61-788-Act1t URA3 Cen | 5 |
| pDA257 | Act1p-GFP-bud6-11-479-Act1t URA3 Cen | 31 |
| pBH432 | Act1p-GFP-Bfa1-Act1t URA3 Cen | This study |
| pBH442 | Act1p-GFP-Bub2-Act1t URA3 Cen | This study |
| pBH444 | Act1p-GFP-Tem1-Act1t URA3 Cen | This study |
| pBH448 | Act1p-GFP-Lte1-Act1t URA3 Cen | This study |
| pFA6a-kanMX6 | Kanr cassette P TEF T TEF | M. Longtine |
| pBJ1351 | TUB1-GFP::LEU2 | 57 |
| pBJ1517 | GAL1-BUD6 | 64 |
Plasmids pBH432, -442, -444, and -448 were created by inserting PCR-amplified coding regions of BFA1, BUB2, TEM1, and LTE1, respectively, into plasmid pTD125 to generate green fluorescent protein (GFP) fusion genes under control of the ACT1 promoter.
Microscopy.
For α-factor analysis, AHY5 was grown to ∼1 × 107 cells/ml. Cells were then diluted to 2 × 106 cells/ml, and α-factor (Sigma, St. Louis, MO) was added to a final concentration of 5 μg/ml (1 mg/ml stock in water). Cells were incubated for ∼3 h (as opposed to the usual ∼105 min) to ensure spindle pole body outer-plaque removal (2). To depolymerize microtubules, FY23x86 cells were treated with 15 μg/ml nocodazole (from a 2-mg/ml stock in dimethyl sulfoxide; Sigma, St. Louis, MO) for up to 135 min at 25°C. For DAPI (4′,6′-diamidino-2-phenylindole) staining other than that described below, cells were fixed using 70% ethanol and incubated in 0.1 μg/ml DAPI for 5 min prior to microscopic analysis.
For Cdc14-GFP experiments, wild-type and bud6Δ strains were treated with 12 μg/ml nocodazole at 25°C. Cells were examined directly from culture or fixed with formaldehyde, washed, and resuspended in glycerol mount containing 50 ng/ml DAPI to visualize DNA. The integrated Cdc14-GFP fusion construct was created by PCR amplification of the C-terminal fusion junction and downstream G418 resistance marker from strain YTC240 (10) and using the resulting PCR product to transform FY23x86 and DAY101x102. Resulting transformants were sporulated, and tetrads were dissected to generate wild-type and bud6Δ haploid strains (BHY444 and BHY443) that have GFP-Cdc14p as their sole source of Cdc14p.
For time-lapse analysis of GFP-bud6-1p SPB mobility, wild-type (FY23x86) or bud6Δ (DAY101x102) homozygous diploid cells carrying pDA257 were grown to mid-log phase in SC lacking uracil (for plasmid selection). Approximately 10 μl of culture was placed on a slide coated with SC in 25% gelatin. A coverslip was placed on top, and cells were observed for GFP fluorescence at ∼15- to 30-s intervals. Distances from SPB spots to the outer boundary of the GFP-bud6-1p neck rings were measured in pixels from the Openlab images, with positive values denoting measurements on the mother side of the neck rings and negative values on the daughter side. An arbitrary value of ±5 pixels (∼0.33 μm) was chosen as the cutoff value for a given SPB to be considered “associated” with the GFP-bud6-1p neck ring structures. Time-lapse observations were begun when one of the SPBs fell within the 5-pixel range from the neck rings of medium- to large-budded cells, which were interpreted as preanaphase cells. Measurements from a given cell were discarded if the inter-SPB distance appeared to be consistently increasing (i.e., the cell was presumed to be entering anaphase). A neck-crossing event was judged to have occurred when a given SPB went from >5 pixels from the neck rings on one side of the neck to >5 pixels on the other side of the neck (e.g., migration of an SPB from daughter to mother cell).
An Axioskop 2 MOT (Carl Zeiss, Germany) with a Plan-APOCHROMAT 100× objective was used for microscopic analysis. Cells were grown to mid-log phase for analysis by epifluorescence using standard fluorescein isothiocyanate, GFP, and cyan fluorescent protein filter sets and by differential interference contrast. Images were captured with an ORCA-ER digital camera (Hamamatsu Photonics, Japan) using Openlab software (Improvision, England). Adobe Photoshop (Adobe Systems, San Jose, CA) was used for final adjustments of the captured images.
Assays to determine the contributions of Arp1p, Bud6p, and Kar9p to spindle position.
Growth rates of YJC3677, -3681, -3685, -3689, -3693, -3697, -3701, and -3705 (Table 1) were determined by measuring the increase in optical density at 600 nm of asynchronous cultures of four independent segregants of each genotype grown in liquid YPD medium at 30°C. Multinucleate cells and mispositioned spindles (late anaphase spindles in the mother) were scored by observing GFP-Tub1 fluorescence in two independent segregants of each genotype. For BUD6 overexpression, arp1Δ GFP-TUB1 cells (strain YJC3681) were transformed with plasmid pBJ1517 (GAL1-BUD6) and grown in rich galactose or glucose media overnight before scoring spindle position in asynchronous cultures. For these experiments, still images were collected on an Olympus IX70 fluorescence microscope (Melville, NY) with a 100×/1.4 oil immersion objective lens and a Coolsnap HQ camera (Roper Scientific, Duluth, GA). Images were acquired using QED Imaging software (Pittsburgh, PA) and analyzed using Image J (W. Rasband, NIH; http://rsb.info.nih.gov/ij/).
RESULTS
GFP-bud6-1p colocalizes with Spc42p-ECFP.
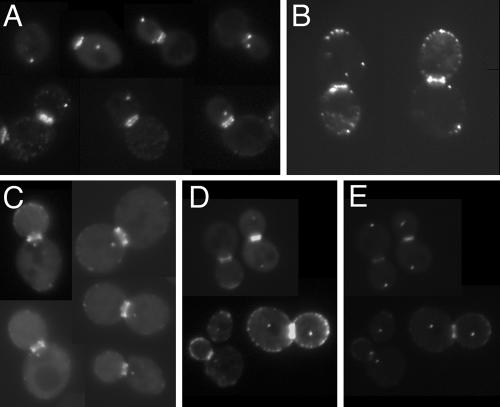
Previous investigations of the intracellular localization of Bud6p involved indirect immunofluorescence microscopy using purified anti-Bud6p antibodies on wild-type cells or by direct visualization of GFP-Bud6p expressed from a plasmid and under control of the ACT1 promoter, which provides a moderate level of constitutive expression (5, 51). Both methods indicated that Bud6p is present at the incipient bud site, at the tips of growing buds, and as a pair of rings at the mother-bud junction, consistent with its recognized functions. Our analysis of yeast cells containing GFP-BUD6 integrated at the URA3 locus and under control of the ACT1 promoter (AHY26; Materials and Methods) revealed that, in addition to the previously reported Bud6p distribution, GFP-Bud6p is present as one or two noncortical spots reminiscent of yeast SPB staining (Fig. 1A). We observed comparable localization patterns with a plasmid-borne full-length GFP-Bud6p fusion construct (pDAb204; Fig. 1B) and with a truncated form of Bud6p (referred to as bud6-1p) that expresses amino acids 1 to 479 (pDA257; Fig. 1C) (31). To determine whether these spots colocalize with a known SPB component, we transformed pDAb204 and pDA257 into cells harboring genome-integrated SPC42-ECFP (Materials and Methods). Spc42p is a component of the SPB central plaque, a substructure of the SPB embedded within the nuclear envelope (24). Both full-length and truncated GFP-Bud6p colocalize with Spc42p-ECFP in unbudded, small-budded, and large-budded cells, suggesting that both fusion proteins are present at the SPBs throughout the yeast cell cycle (Fig. 1D and E and data not shown). Although some GFP-Bud6p signal bleeds through the CFP filter set, no signal from Spc42-ECFP bleeds through the GFP filter set, and unfused GFP shows no SPB localization (data not shown). Thus, we are confident that Bud6p, in addition to its neck and cortical spot localization, associates with the SPBs. Because we consistently observed less background fluorescence with the construct pDA257 (GFP-bud6-1p) in comparison to pDAb204 (GFP-Bud6p) and because GFP-bud6-1p does not complement the growth, actin organization, or nuclear positioning defects of a bud6Δ allele (31; our unpublished observations), pDA257 was used for most of our subsequent analyses. Use of this nonfunctional bud6p fusion has allowed us to examine Bud6p localization in the absence of normal Bud6p function. We have observed no adverse dominant effects of expressing bud6-1p in BUD6 genetic backgrounds.
FIG. 1.
GFP-Bud6p localizes at SPB-like dots. (A) Diploid wild-type yeast strain AHY26 (Table 1) containing integrated GFP-BUD6 shows staining at one or two SPB-like dots, in addition to the incipient bud site, tips of small buds, and mother-bud neck region. (B and C) Diploid wild-type yeast strain FY23x86 (Table 1) variants expressing plasmid-borne (B) full-length GFP-Bud6p from plasmid pDAb204 or (C) truncated GFP-bud6-1p from plasmid pDA257 show similar distributions of GFP staining. (D and E) GFP-bud6-1p expressed from pDA257 in AHY5 shows colocalization of GFP-bud6-1 “SPB” spots (D) with the SPB component Spc42p (E).
GFP-bud6-1p is present at the SPB independent of dynein and microtubules.
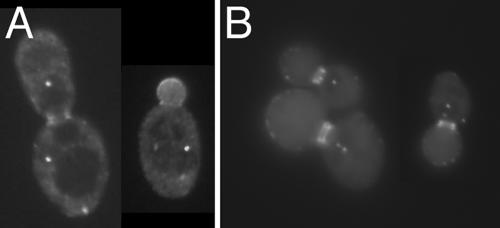
Previous reports that Bud6p functions to aid in microtubule capture at the bud cortex and the neck (25, 51, 52) led us to investigate the possibility that Bud6p may be transported along microtubules in a minus-end-directed movement to the SPB. We first tested whether the minus-end-directed microtubule motor dynein plays a role in carrying Bud6p to the SPB. Analysis of a dyn1 deletion strain (PY23643x3297) containing pDA257 demonstrated that SPB localization of GFP-bud6-1p is not impaired in diploid cells lacking dynein (Fig. 2A). To determine if Bud6p is transported to the SPB via microtubules, we treated yeast cells harboring pDA257 with nocodazole so as to depolymerize microtubules. Cells were observed at various intervals up to 135 min. SPB localization of GFP-bud6-1p was never compromised at the observed time points (Fig. 2B shows 105 min after nocodazole treatment). This finding suggests that microtubules are not required to maintain GFP-bud6-1p at the SPB.
FIG. 2.
GFP-bud6-1p stains the SPB in the absence of dynein or microtubules. (A) Homozygous dynein deletion strain PY23643x3297 (Table 1) expressing GFP-bud6-1p from pDA257. (B) Wild-type diploid strain FY23x86 carrying pDA257 was treated with nocodazole, and GFP-bud6-1p fluorescence was observed at various times; the image shown is of cells after 105 min of exposure to nocodazole, at which point 94% of cells had medium to large buds.
Localization of GFP-bud6-1p at the SPB requires an intact outer-plaque structure.
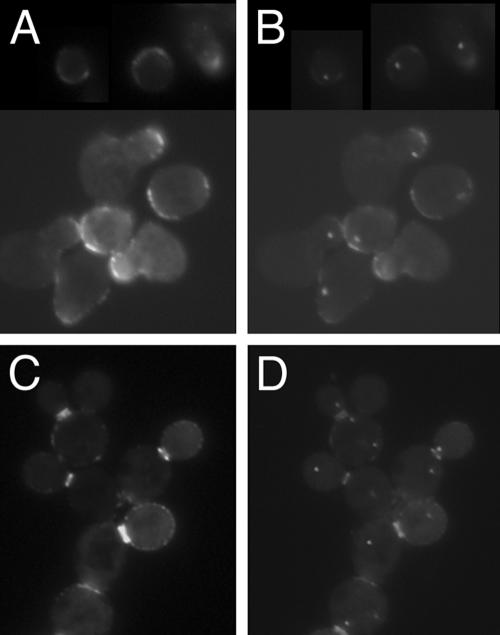
Considering the role of Bud6p in such cytoplasmic events as cell polarity and microtubule capture at the cell periphery (5, 25, 51, 52), we reasoned it more likely that Bud6p is present at or near the SPB outer plaque, the SPB substructure that is oriented towards the cytoplasm and nucleates astral microtubules. Nud1p is an outer-plaque component located directly subjacent to the gamma-tubulin complex-binding protein Spc72p (24). The nud1-44 allele causes a conditional arrest characterized by cells bearing SPBs with an aberrant outer plaque that is incapable of nucleating cytoplasmic microtubule arrays (2). We transformed pDA257 into the nud1-44 mutant (IAY520) to learn whether Nud1p and the SPB components that reside external to Nud1p in the outer plaque (including Spc72p and the gamma-tubulin complex proteins Spc97p, Spc98p, and Tub4p) are necessary for GFP-bud6-1p localization to the spindle poles. After 4 h at restrictive temperature, many nud1-44 cells were found to contain GFP-bud6-1p staining of the SPB and this staining was similar to that of a congenic wild-type strain (IAY522) under the same conditions (Fig. 3). (Note that it was difficult to observe SPB-like GFP-bud6-1p dots in every cell due to a high background, although GFP-bud6-1p staining at the bud tip and neck region remained clear.) These findings imply that GFP-bud6-1p localization is not affected by expression of the nud1-44 allele.
FIG. 3.
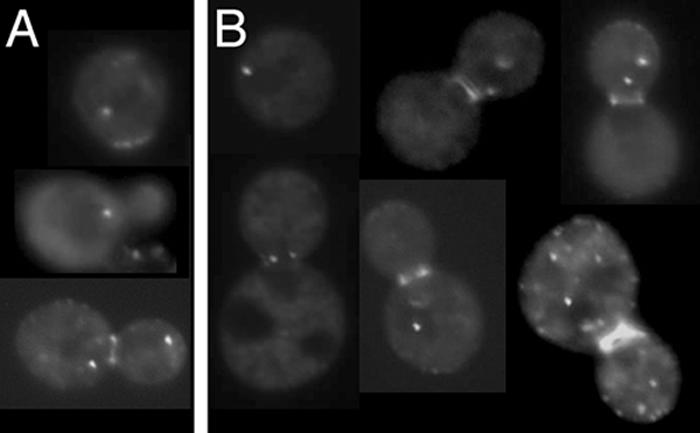
GFP-bud6-1p is present as SPB-like dots in nud1-44 cells. Both (A) wild-type NUD1 (IAY522) and (B) mutant nud1-44 (IAY520) (Table 1) cells containing pDA257 maintain GFP-bud6-1p at the SPB after 4 h at the nud1-44 restrictive temperature of 37°C.
Based on our findings with nud1-44, we decided to test whether the SPB outer plaque as a whole is important for accumulation of GFP-bud6-1p at the SPBs. We treated an SPC42-ECFP strain (AHY5) containing pDA257 with α-factor for ∼3 h, a procedure previously shown to cause complete removal of the outer plaque without loss of the central plaque component Spc42p (2). We discovered that upon prolonged α-factor exposure, GFP-bud6-1p does not colocalize with Spc42p-ECFP (Fig. 4A and B). Prior to α-factor exposure, 92% of cells contained GFP-bud6-1p staining at Spc42p-ECFP spots (not shown), whereas after a 3-h exposure to α-factor, only 18% of cells maintained such colocalization (n ≥ 100 cells). Moreover, within this 18% pool, the GFP-bud6-1p SPB staining was significantly dimmer, suggesting that residual staining may be due to incomplete outer-plaque removal in these cells. GFP-bud6-1p does concentrate at mating projections, indicating that α-factor does not have a general effect on GFP-bud6-1p localization. Furthermore, release from α-factor brings GFP-bud6-1p back to the spindle pole (data not shown), consistent with a reported restoration of the multilaminar structure of the SPB under similar conditions (2). These results suggest that GFP-bud6-1p is present only at the cytoplasmic side of the SPB, as the inner plaque (on the nucleoplasmic side of the SPB) is not altered upon extensive α-factor treatment.
FIG. 4.
The SPB outer plaque is required for localization of Bud6p to the SPB. (A and B) Strain AHY5 carrying pDA257 was treated with α-factor for 3 h as described in Materials and Methods, and cells were observed for GFP-bud6-1p (A) and Spc42p-ECFP (B). (C and D) The cnm67 mutant strain AHY13 carrying pDA257 was observed for GFP-bud6-1p (C) and Spc42p-ECFP (D). Note that Spc42p-ECFP is still able to stain the SPB, as the central plaque is not affected in cnm67 null cells.
In the absence of the SPB outer-plaque component Cnm67p, cells harbor SPBs that lack an outer-plaque structure (8). Such cells display a severe nuclear segregation defect but are viable due to their ability to nucleate astral microtubules from the half-bridge structure, a region of the nuclear envelope adjacent to the disk-shaped portion of the SPB (24). To assess the potential role of Cnm67p in localizing GFP-bud6-1p at the SPB, we constructed a cnm67Δ strain expressing Spc42p-ECFP (AHY13; Materials and Methods) and containing pDA257. GFP-bud6-1p failed to colocalize with Spc42p-ECFP in most cnm67 cells (Fig. 4C and D), although 5% of cells (n > 100) did show GFP-bud6-1p SPB localization. Our observation of more than one or two dots of Spc42p-ECFP staining per cell is consistent with the reported nuclear migration defects of a cnm67 null mutant, causing cells to accumulate multiple SPBs (8). In addition, we noticed that even in the absence of Cnm67p, GFP-bud6-1p is able to mark the incipient bud site and appears as two rings at the mother-bud neck. Thus, we conclude that Cnm67p is essential for Bud6p localization to SPBs but not for its cortical or neck localization.
GFP-bud6-1p behavior in wild-type and bud6Δ cells.
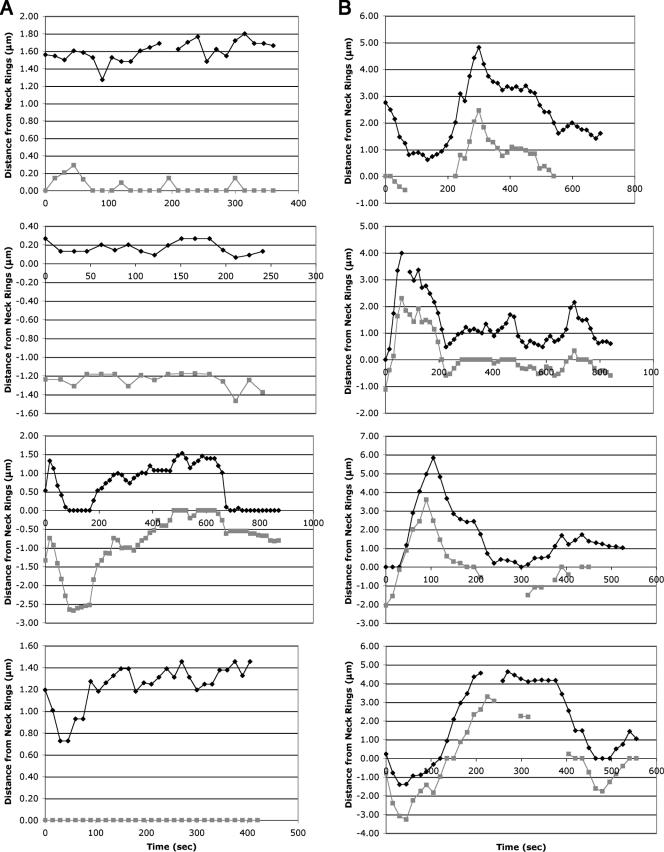
Others have indicated a role for Bud6p in maintaining microtubule contact at the cell cortex and in properly positioning the nucleus during mitosis (25, 26, 41, 51, 52, 62). We similarly examined the effect of bud6 deletion on truncated bud6-1p localization and SPB movement. Our initial observations of SPB movement in a bud6Δ strain suggested excessive spindle oscillation, as was similarly observed previously (51, 62). In order to more carefully characterize this SPB behavior in bud6Δ cells, we followed GFP-bud6-1p localization over time in asynchronous wild-type and bud6Δ/bud6Δ diploid strains. The distances of the SPBs from the GFP-bud6-1p-labeled neck rings were measured at 15-s intervals and tabulated to determine the time a given SPB remains “associated” with the mother-bud neck (see Materials and Methods for measuring and scoring criteria). To avoid potential bias, only large-budded cells that had properly positioned spindles at the beginning of the experiment were analyzed. In wild-type cells (Fig. 5A and see Table S1 and Movies S2a and S2b in the supplemental material), one of the two SPBs is generally found associated with the neck (42.7% of the total SPB time recorded; note that 50% is the maximum attainable value if only one of the two SPBs is associated with the neck at one time). The SPBs of mutants defective for Bud6p function, however, spend considerably less time associated with the neck (31.3% of total SPB time recorded; Fig. 5B and see Table S2 and Movies S2c and S2d in the supplemental material) and more frequently oscillate between mother and daughter (“neck crossings”). Furthermore, the average duration of association of a given SPB with the neck region is considerably less for bud6Δ cells than for the wild type (47 s versus 135 s for SPBs whose neck association was recorded from start to finish or 67 s versus 229 s for all SPB recordings, noting that some of these observations began and/or ended with the SPBs associated with the neck). Overall, these data indicate that SPBs from bud6Δ mutants are less tightly associated with neck structures, resulting in more frequent preanaphase nuclear oscillations and potentially aberrant nuclear positioning.
FIG. 5.
Graphical representations of SPB movements in wild-type and bud6Δ/bud6Δ diploid cells. The distances of GFP-bud6-1p spots from neck ring structures were measured at 15-s intervals in representative cells of (A) the wild type (FY23x86) or (B) bud6Δ/bud6Δ (DAY101x102) diploids carrying plasmid pDA257. Positive and negative values denote positions on the mother and daughter sides of the neck, respectively; gray and black lines distinguish the two SPBs.
Bud6p does not contribute to spindle entry into the bud.
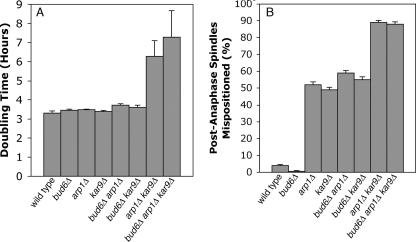
Previous studies have established that the Bud6p-microtubule interaction at the cell cortex is important to bring the short spindle into alignment with the mother-bud axis (6), and the above results suggest that Bud6p-neck interactions are important for maintaining nuclear position at the neck. We next investigated whether the microtubule-Bud6p interaction helps to pull the spindle through the mother-bud neck, as the dynein and Kar9p pathways have been shown to do (1, 52). This question has not been specifically addressed previously. First, we confirmed that a bud6 mutant has a spindle orientation defect, as previously shown (52; data not shown). To examine a possible role for Bud6p related to the Kar9p and dynein pathways, we tested for a synthetic effect on growth of mutants lacking BUD6 in combination with null alleles of the dynein and Kar9p pathways. Cells lacking essential components of the Kar9p and dynein pathways move the spindle into the neck poorly and are very sick (1). Loss of BUD6 had no effect on growth of kar9Δ, arp1Δ (ARP1 encodes a component of the dynactin complex), or arp1Δ kar9Δ mutants (Fig. 6A). These data indicate that Bud6p is not an essential component of either spindle-positioning pathway.
FIG. 6.
The role of Bud6p in pulling the spindle through the neck. (A) A bud6Δ mutation does not exacerbate the growth defects of kar9 or arp1 (dynactin) mutants, indicating that Bud6p does not have an important role in either Kar9p or dynein-mediated spindle position; doubling times of the indicated strains are expressed in hours. (B) A bud6 mutation does not increase the frequency with which cells misposition their spindles in wild-type, arp1, or kar9 mutant cells (expressed as percent anaphase spindles in the mother). The strains used for these experiments were YJC3677, -3681, -3685, -3689, -3693, -3697, -3701, and -3705 (Table 1).
To determine more directly if Bud6p has a role in moving mitotic spindles through the neck, we counted how often late-anaphase spindles were seen in the mother in single, double, and triple mutants. Wild-type cells and cells lacking only BUD6 had no late-anaphase spindles in the mother (Fig. 6B). Half of the arp1Δ or kar9Δ cells had long anaphase spindles in the mother, and loss of BUD6 did not increase the fraction of mispositioned spindles observed in arp1Δ or kar9Δ cells. Therefore, the Bud6p-microtubule interaction does not appear to contribute to moving the spindle from the mother into the neck under these conditions. Instead, we propose that the most important consequence of this interaction is to sense or stabilize spindle position.
Overexpression of Bud6p enhances SPB-neck interactions.
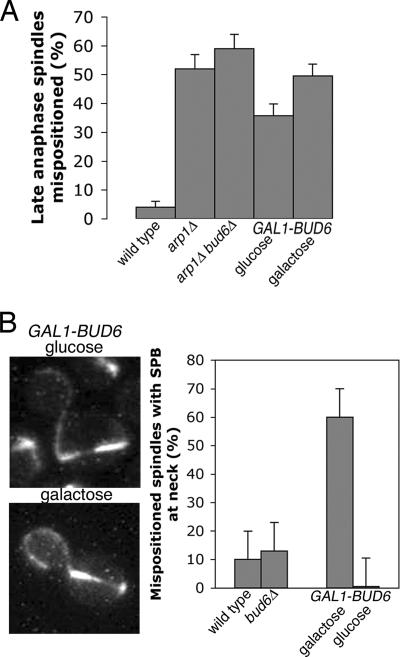
To further investigate the role of Bud6p in spindle position and SPB-neck interactions, we overexpressed Bud6p in a dynein pathway mutant (arp1Δ). If Bud6p has a role in pulling the spindle through the mother-bud neck, then overexpression of BUD6 should partially suppress a dynein pathway mutant. When we overexpressed BUD6 from the GAL1 promoter, the frequency with which spindles entered the neck did not change, confirming that Bud6p does not pull the spindle through the neck (Fig. 7A).
FIG. 7.
The role of Bud6 in spindle alignment and SPB-neck interaction. (A) Overexpression of BUD6 does not suppress the spindle position defect of a dynactin (arp1Δ) mutant. The percentage of mispositioned late-anaphase spindles were counted (expressed as percent long spindles in the mother) in the wild-type, arp1Δ (YJC3681), and arp1Δ bud6Δ (YJC3697) strains and in an arp1Δ strain carrying a galactose-inducible BUD6 gene (YJC4035) grown in glucose and galactose. (B) Overexpression of BUD6 in an arp1Δ mutant restores spindle alignment relative to the mother-bud axis, and the old SPB is at the neck. (Left) Representative YJC4035 cells grown in glucose or galactose. (Right) Percent anaphase spindles in the mother with a SPB at the neck measured in wild-type and YJC3689 (bud6Δ) cells and in YJC4035 cells (arp1Δ GAL1-BUD6) grown in glucose and galactose.
Overexpression of BUD6 did enhance the frequency of SPB interactions with the neck (Fig. 7B). In arp1Δ cells, 90% of late-anaphase spindles in the mother were nearly perpendicular to the mother-bud axis, with neither spindle pole body in the vicinity of the neck. After galactose induction of GAL1-BUD6, one SPB touched the neck in 60% of cells with late-anaphase spindles in the mother. In glucose, no SPBs touched the neck. These results suggest that an important function of Bud6p is to align the mitotic spindle relative to the mother-bud axis, presumably due to Bud6p-mediated interactions between the SPBs and the bud neck.
Loss of Bud6p does not perturb the spindle position checkpoint.
The aberrant spindle oscillations observed in bud6Δ cells suggested possible defects in regulation of the mitotic exit network (MEN) or the spindle position checkpoint (33). Key regulators of MEN include the small GTPase Tem1p; its two-component GTPase-activating protein (GAP) comprised of Bub2p and Bfa1p; and the Tem1p guanine nucleotide exchange factor, Lte1p. Tem1p, Bub2p, and Bfa1p are localized primarily to the spindle poles, likely maintaining Tem1p in the GDP-bound “off” state until it makes contact with Lte1p, which is localized at the periphery of the growing bud. Exchange of GDP for GTP on Tem1p is thought to initiate MEN signaling, although this remains controversial (14, 21). To determine if Bud6p participates in maintaining Bub2p, Bfa1p, or Tem1p at the SPBs or in bringing Lte1p to the cell periphery, we expressed GFP fusions of these proteins in wild-type and bud6Δ strains. In each case, we found that loss of Bud6p did not significantly alter the localization of these proteins (see Fig. S1 in the supplemental material and see reference 30 for Bud6p independence of Lte1p localization). We conclude that loss of Bud6p does not significantly impact these components of the MEN signaling pathway.
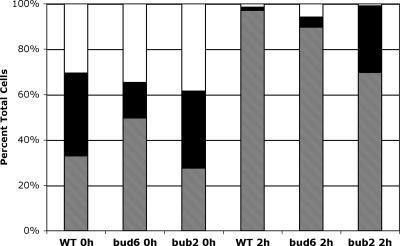
Another key regulator of mitotic exit is the regulatory phosphatase Cdc14p. This protein is usually sequestered in the nucleolus until MEN activation causes its release, allowing it to act on targets that promote cyclin degradation and exit from mitosis. One hallmark of defects in the spindle position checkpoint is the premature release of Cdc14p from the nucleolus upon microtubule depolymerization; i.e., in the absence of an active checkpoint, these cells will release Cdc14p and prematurely attempt to complete mitosis. Treatment of bud6Δ cells with nocodazole caused only minor release of GFP-Cdc14p from the nucleolus (Fig. 8), suggesting that this aspect of the spindle position checkpoint is largely unperturbed in the absence of Bud6p. As a control, Cdc14p was released in a significant proportion of nocodazole-treated bub2Δ cells, consistent with a loss of checkpoint control. Notably, untreated bud6Δ cells spent proportionately less time in anaphase, as judged by Cdc14-GFP localization. The higher proportion of uninucleate bud6Δ cells (Fig. 8, single nucleolar staining in untreated cells) suggests a possible delay prior to mitosis or at the metaphase-to-anaphase transition; we are currently investigating this aspect of the bud6Δ phenotype.
FIG. 8.
bud6Δ cells are not defective for the spindle position checkpoint. Cdc14-GFP localization was followed in wild type (BHY444), bud6Δ (BHY443), and bub2Δ (BHY482) haploid cells before (0 h) or 2 h after addition of nocodazole. Nucleolar Cdc14-GFP was scored as a single cluster in mother or daughter cells (hatched) versus two or more clusters in mother and daughter cells (white); “diffuse” cells (black) had no obvious nucleolar staining. n > 150 cells for each time point.
DISCUSSION
In this report, we provide the first evidence that Bud6p/Aip3p localizes to the SPB in Saccharomyces cerevisiae. As for bud and neck localization of Bud6p, SPB localization requires only the N-terminal 479 amino acids of Bud6p (31) and does not require the C-terminal effector domain known to be required for actin (5) and polarisome interactions (20, 55) and for activation of the formin Bni1p (42). We demonstrated that this localization is independent of the minus-end-directed motor dynein (Fig. 2A) and that maintenance of GFP-bud6-1p at the SPBs does not require intact microtubules (Fig. 2B). We further showed that the core SPB outer plaque component Cnm67p is required for localizing GFP-bud6-1p to the SPB (Fig. 4). In addition, we observed that in the absence of Bud6p the preanaphase nucleus shows significant oscillations and a marked inability to stably position itself in the vicinity of the mother-bud neck (Fig. 5B and see Table S2 in the supplemental material); similar observations have been made by others (51, 62). Unstable nuclear positioning in bud6Δ cells appears to correlate with an inability to form interactions between either of the SPBs and the neck.
Given Bud6p's diverse roles in regulating cell polarity and spindle positioning, its overlapping localization with most of the proteins of the MEN, and the propensity of bud6Δ cells to generate multinucleate cells (5), it is tempting to theorize that Bud6p could be helping to coordinate cell cycle events at the SPB by affecting the activity of MEN components. One mechanism that Bud6p could utilize to affect Tem1p activity would be to regulate the localization of the Bub2p/Bfa1p GAP, but we have found no loss of Bub2p or Bfa1p from the SPB in bud6Δ strains (see Fig. S1 in the supplemental material). Alternatively, Bud6p could modulate the activity of the Bub2p/Bfa1p GAP either directly or through Cdc5p, a polo-like kinase known to inhibit Bub2p/Bfa1p by phosphorylation (22). Interestingly, Cdc5p colocalizes with Bud6p at the bud neck and the SPB (56), but bud6Δ cells are not defective for Cdc5p localization (data not shown). An additional possibility is that Bud6p could directly affect the activity of the Tem1p GTPase or affect the activity of downstream MEN components that colocalize to the neck and/or SPB. That we do not see additive effects of combining bud6Δ and bub2Δ, bfa1Δ, or lte1Δ mutations suggests that loss of Bud6p does not tip the balance of the Tem1p GTPase cycle sufficiently to either block mitosis or cause premature activation of the MEN (e.g., by failing to inactivate or activate, respectively, Bub2p/Bfa1p). Arguing against an activating role for Bud6p in the spindle-positioning checkpoint (inhibitory to MEN) are observations by us (5) and others (51) that bud6Δ cells transit late stages of the cell cycle more slowly than wild-type cells and that Cdc14p is not released from the nucleolus upon nocodazole treatment of bud6Δ cells (Fig. 8), though we cannot rule out the possibility that nocodazole treatment activates additional checkpoint controls (e.g., spindle-assembly or “no-cut” checkpoints [33, 43]), thereby masking spindle position checkpoint defects. The lack of elongated late-anaphase spindles in bud6Δ mother cells (Fig. 6B) also argues against Bud6p playing a significant role in the spindle position checkpoint.
Current models suggest that entry of a SPB into the daughter cell is responsible for stimulating mitotic exit (e.g., see reference 26). Our results indicate that, at least in our strain backgrounds, this is likely untrue. We have found that either SPB may be associated with the neck prior to anaphase: in cases where SPBnew (mother) is associated with the neck, SPBold (daughter) spends a significant amount of time in the bud without concomitant entry into anaphase (see Fig. 5A). A more likely scenario is one in which cumulative time of association of either SPB with neck components determines mitotic progression, either through inactivation of spindle-positioning checkpoint controls or through activation of anaphase-promoting complex and/or MEN pathways. It is possible that SPBold is more competent to promote activation, but our results suggest that SPBnew-neck association can also function in this capacity. This is entirely consistent with the recent findings of Magidson et al. (38) that either Schizosaccharomyces pombe SPB can promote cytokinesis. That bud6Δ mutants are delayed in mitotic progression is also consistent with a role for SPB-neck contact promoting mitotic progression. One plausible explanation for these observations is that progression into anaphase is promoted by bringing SPB components, such as Bfa1p/Bub2p, to the mother-bud neck, where they may be acted upon by the Cdc5p kinase, resulting in activation of the Tem1p GTPase. This could work in concert with the additional activation of Tem1p by the (nonessential) exchange factor Lte1p at the daughter cell cortex. Notably, the contribution by Lte1p would make the daughter-bound SPBold the apparent major, but not sole, contributor to MEN activation.
Bud6p's role in stabilizing preanaphase spindle position could be purely mechanical. In a series of studies, Segal, Bloom, and coworkers (25, 51, 52, 62) have closely monitored microtubule-cortex interactions in bud6, bni1, and kar9 strains and have proposed that Bud6p-dependent sequential interactions of astral microtubules with the bud cortex and bud neck facilitate spindle orientation and subsequent stabilization of the nucleus within the neck. Our observations of unstable nuclear positioning in bud6Δ cells are in agreement with similar results from these researchers (51, 62). While providing elegant analyses of the relative contributions of Bud6p and Kar9p to microtubule interactions at the cortex, we feel the previously reported data and proposed models do not fully explain Bud6p's role in retaining the preanaphase spindle at the neck. For example, it is difficult to imagine how microtubule-neck interactions could be simultaneously maintained by both SPBs or established rapidly enough to prevent either SPB from escaping the neck in a nucleus that is rapidly oscillating through the neck. Indeed, Jacobs et al. (27) have demonstrated that although microtubules are essential for establishing nuclear orientation, they are not required for retaining the nucleus at the neck once such positioning has been established.
Our discovery of Bud6p SPB localization, our analysis of GFP-bud6-1p behavior in wild-type and bud6Δ strains (Fig. 5 and see Tables S1 and S2 and Movies S2a to S2d in the supplemental material), and our observation that excess Bud6p can promote SPB-neck interactions (Fig. 7B) support a more direct role for Bud6p in keeping the nucleus bound at the neck via Bud6p-mediated SPB-neck interactions. The C-terminal region of Bud6p contains an extensive heptad repeat region predicted to form coiled-coil interactions. Consequently, the inability of GFP-bud6-1p, which lacks this C-terminal region, to complement spindle-positioning defects might arise from its inability to interact with other heptad repeat proteins, including substrates at the mother-bud neck or SPB. Interestingly, several components of the SPB outer plaque, including Cnm67p and Spc72p, contain extensive heptad repeat domains (24) and could be targets for interaction with Bud6p present at the mother-bud neck, although we have been unable to detect an interaction between Bud6p and these or other (Spc97p, Spc98p, or Spc110p) SPB components (unpublished data). The ability of bud6-1p to localize at the SPB could be due to a smaller heptad repeat region (which shows some similarity to the Cnm67p heptad repeat domain) present in the N-terminal domain or to non-coiled-coil interactions. Similarly, four out of five septins, which are major structural components and polarity determinants of the mother-bud neck, contain C-terminal heptad repeat domains and could serve as sites of interaction with SPB-bound Bud6p. It is also possible that homo-oligomeric interactions between Bud6p molecules (5) of the SPB and neck may contribute to preanaphase nuclear stabilization. Finally, it is possible that Bud6p plays both mechanical and regulatory roles at the SPB: e.g., by utilizing neck-bound Bud6p-microtubule interactions to bring either SPB close enough to stabilize the preanaphase nucleus at the neck via SPB-bound Bud6p.
The presence of Bud6p at the neck and SPBs, coupled with aberrant nuclear movement in bud6Δ cells, strongly suggests a role for Bud6p in providing quality control to preanaphase nuclear positioning. Based on the prominent preanaphase association of one SPB with the neck and the lack of “neck-crossing” events in wild-type cells, it is likely that Bud6p-mediated associations provide a barrier to block excessive movement of SPBs past components of the mother-bud neck. Perhaps more importantly, we believe that Bud6p also plays an important role in stimulating mitotic progression by bringing regulatory components at the SPBs and bud neck into close proximity with one another. Future studies will help to determine the nature of these interactions and the identity of Bud6p's binding partners that contribute to this novel pathway.
Supplementary Material
Acknowledgments
We thank the Kilmartin and Pellman laboratories for kindly providing constructs. We thank Tatiana Yuzyuk, Blaine Bettinger, and Sue Viggiano for their helpful suggestions and technical assistance.
This research was funded by National Institutes of Health grants GM056189 to D.C.A. and GM69895 to J.A.C.
Footnotes
Published ahead of print on 6 April 2007.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Adames, N. R., and J. A. Cooper. 2000. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 149:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, I. R., and J. V. Kilmartin. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145:809-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amberg, D. C., D. Botstein, and E. M. Beasley. 1995. Precise gene disruption in Saccharomyces cerevisiae by double fusion polymerase chain reaction. Yeast 11:1275-1280. [DOI] [PubMed] [Google Scholar]
- 4.Amberg, D. C., D. J. Burke, and J. N. Strathern. 2005. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Amberg, D. C., J. E. Zahner, J. W. Mulholland, J. R. Pringle, and D. Botstein. 1997. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell 8:729-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bardin, A. J., R. Visintin, and A. Amon. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102:21-31. [DOI] [PubMed] [Google Scholar]
- 7.Beach, D. L., J. Thibodeaux, P. Maddox, E. Yeh, and K. Bloom. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497-1506. [DOI] [PubMed] [Google Scholar]
- 8.Brachat, A., J. V. Kilmartin, A. Wach, and P. Philippsen. 1998. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol. Biol. Cell 9:977-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byers, B., and L. Goetsch. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, T., J. Aulds, T. Gill, M. Cerio, and M. E. Schmitt. 2002. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics 161:1029-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao, L. G., and Y. L. Wang. 1996. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol. Biol. Cell 7:225-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carminati, J. L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenn, A., and S. K. McConnell. 1995. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell 82:631-641. [DOI] [PubMed] [Google Scholar]
- 14.Cooper, J. A., and S. A. Nelson. 2006. Checkpoint control of mitotic exit—do budding yeast mind the GAP? J. Cell Biol. 172:331-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cottingham, F. R., L. Gheber, D. L. Miller, and M. A. Hoyt. 1999. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147:335-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cottingham, F. R., and M. A. Hoyt. 1997. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J. Cell Biol. 138:1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeZwaan, T. M., E. Ellingson, D. Pellman, and D. M. Roof. 1997. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J. Cell Biol. 138:1023-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drubin, D. G., and W. J. Nelson. 1996. Origins of cell polarity. Cell 84:335-344. [DOI] [PubMed] [Google Scholar]
- 19.Eshel, D., L. A. Urrestarazu, S. Vissers, J. C. Jauniaux, J. C. van Vliet-Reedijk, R. J. Planta, and I. R. Gibbons. 1993. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA 90:11172-11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evangelista, M., K. Blundell, M. S. Longtine, C. J. Chow, N. Adames, J. R. Pringle, M. Peter, and C. Boone. 1997. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276:118-122. [DOI] [PubMed] [Google Scholar]
- 21.Fraschini, R., C. D'Ambrosio, M. Venturetti, G. Lucchini, and S. Piatti. 2006. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 172:335-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geymonat, M., A. Spanos, P. A. Walker, L. H. Johnston, and S. G. Sedgwick. 2003. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 278:14591-14594. [DOI] [PubMed] [Google Scholar]
- 23.Gonczy, P., and A. A. Hyman. 1996. Cortical domains and the mechanisms of asymmetric cell division. Trends Cell Biol. 6:382-387. [DOI] [PubMed] [Google Scholar]
- 24.Helfant, A. H. 2002. Composition of the spindle pole body of Saccharomyces cerevisiae and the proteins involved in its duplication. Curr. Genet. 40:291-310. [DOI] [PubMed] [Google Scholar]
- 25.Huisman, S. M., O. A. Bales, M. Bertrand, M. F. Smeets, S. I. Reed, and M. Segal. 2004. Differential contribution of Bud6p and Kar9p to microtubule capture and spindle orientation in S. cerevisiae. J. Cell Biol. 167:231-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huisman, S. M., and M. Segal. 2005. Cortical capture of microtubules and spindle polarity in budding yeast—where's the catch? J. Cell Sci. 118:463-471. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs, C. W., A. E. Adams, P. J. Szaniszlo, and J. R. Pringle. 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107:1409-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaquenoud, M., M. P. Gulli, K. Peter, and M. Peter. 1998. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 17:5360-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaquenoud, M., and M. Peter. 2000. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p). Mol. Cell. Biol. 20:6244-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen, S., M. Geymonat, A. L. Johnson, M. Segal, and L. H. Johnston. 2002. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 115:4977-4991. [DOI] [PubMed] [Google Scholar]
- 31.Jin, H., and D. C. Amberg. 2000. The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol. Biol. Cell 11:647-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, L., S. K. Klee, M. Evangelista, C. Boone, and D. Pellman. 1999. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J. Cell Biol. 144:947-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lew, D. J., and D. J. Burke. 2003. The spindle assembly and spindle position checkpoints. Annu. Rev. Genet. 37:251-282. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y. Y., E. Yeh, T. Hays, and K. Bloom. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA 90:10096-10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liakopoulos, D., J. Kusch, S. Grava, J. Vogel, and Y. Barral. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112:561-574. [DOI] [PubMed] [Google Scholar]
- 36.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 37.Maekawa, H., T. Usui, M. Knop, and E. Schiebel. 2003. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 22:438-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magidson, V., F. Chang, and A. Khodjakov. 2006. Regulation of cytokinesis by spindle-pole bodies. Nat. Cell Biol. 8:891-893. [DOI] [PubMed] [Google Scholar]
- 39.McCarthy, E. K., and B. Goldstein. 2006. Asymmetric spindle positioning. Curr. Opin. Cell Biol. 18:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, R. K., K. K. Heller, L. Frisen, D. L. Wallack, D. Loayza, A. E. Gammie, and M. D. Rose. 1998. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol. Biol. Cell 9:2051-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, R. K., D. Matheos, and M. D. Rose. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moseley, J. B., I. Sagot, A. L. Manning, Y. Xu, M. J. Eck, D. Pellman, and B. L. Goode. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell 15:896-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norden, C., M. Mendoza, J. Dobbelaere, C. V. Kotwaliwale, S. Biggins, and Y. Barral. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125:85-98. [DOI] [PubMed] [Google Scholar]
- 44.Palmer, R. E., D. S. Sullivan, T. Huffaker, and D. Koshland. 1992. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 119:583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pringle, J. R., E. Bi, H. A. Harkins, J. E. Zahner, C. De Virgilio, J. Chant, K. Corrado, and H. Fares. 1995. Establishment of cell polarity in yeast. Cold Spring Harbor Symp. Quant. Biol. 60:729-744. [DOI] [PubMed] [Google Scholar]
- 46.Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297:612-615. [DOI] [PubMed] [Google Scholar]
- 47.Pruyne, D. W., D. H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931-1945. [DOI] [PubMed] [Google Scholar]
- 48.Sagot, I., S. K. Klee, and D. Pellman. 2002. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4:42-50. [DOI] [PubMed] [Google Scholar]
- 49.Sagot, I., A. A. Rodal, J. Moseley, B. L. Goode, and D. Pellman. 2002. An actin nucleation mechanism mediated by Bni1 and Profilin. Nat. Cell Biol. 4:626-631. [DOI] [PubMed] [Google Scholar]
- 50.Saunders, W., D. Hornack, V. Lengyel, and C. Deng. 1997. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J. Cell Biol. 137:417-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segal, M., K. Bloom, and S. I. Reed. 2000. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell 11:3689-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal, M., K. Bloom, and S. I. Reed. 2002. Kar9p-independent microtubule capture at Bud6p cortical sites primes spindle polarity before bud emergence in Saccharomyces cerevisiae. Mol. Biol. Cell 13:4141-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segal, M., D. J. Clarke, P. Maddox, E. D. Salmon, K. Bloom, and S. I. Reed. 2000. Coordinated spindle assembly and orientation requires Clb5p-dependent kinase in budding yeast. J. Cell Biol. 148:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaw, S. L., P. Maddox, R. V. Skibbens, E. Yeh, E. D. Salmon, and K. Bloom. 1998. Nuclear and spindle dynamics in budding yeast. Mol. Biol. Cell 9:1627-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheu, Y.-J., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song, S., T. Z. Grenfell, S. Garfield, R. L. Erikson, and K. S. Lee. 2000. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell. Biol. 20:286-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, S., and K. S. Lee. 2001. A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J. Cell Biol. 152:451-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan, D. S., and T. C. Huffaker. 1992. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J. Cell Biol. 119:379-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Theesfeld, C. L., J. E. Irazoqui, K. Bloom, and D. J. Lew. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146:1019-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winey, M., and E. T. O'Toole. 2001. The spindle cycle in budding yeast. Nat. Cell Biol. 3:E23-E27. [DOI] [PubMed] [Google Scholar]
- 61.Yeh, E., R. V. Skibbens, J. W. Cheng, E. D. Salmon, and K. Bloom. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeh, E., C. Yang, E. Chin, P. Maddox, E. D. Salmon, D. J. Lew, and K. Bloom. 2000. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell 11:3949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin, H., D. Pruyne, T. C. Huffaker, and A. Bretscher. 2000. Myosin V orientates the mitotic spindle in yeast. Nature 406:1013-1015. [DOI] [PubMed] [Google Scholar]
- 64.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 293:2101-2105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.