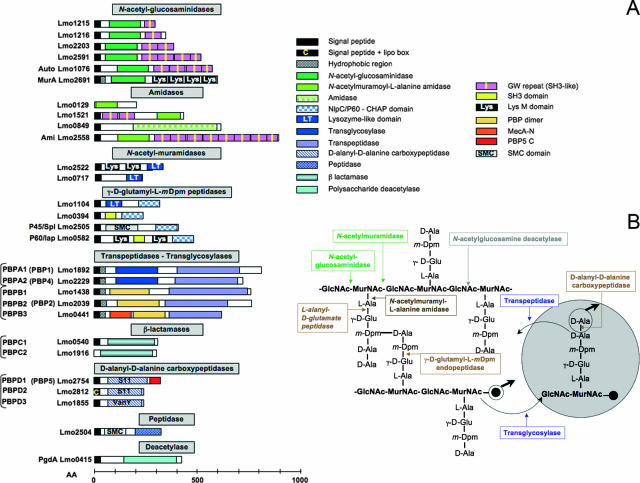
FIG. 3.
Surface proteins predicted to be involved in cell wall metabolism. (A) Schematic representation of L. monocytogenes surface proteins potentially involved in cell wall synthesis, hydrolysis, or modification. AA, amino acids. Most of the enzymatic activities are deduced from domain sequence similarities and require biochemical experimental validation. (B) Cell wall synthetase and hydrolase activities on Listeria peptidoglycan. Glycan chains of GlcNAc and MurNAc are interlinked by direct peptide linkage between the meso-diaminopimelic acid (m-Dpm) residue of one wall peptide and the d-alanyl residue at position 4 of the adjacent wall peptide. A peptidoglycan precursor linked to the undecaprenyl pyrophosphate group (black circle) is represented by a spotted circle. The precursor is incorporated to the preexisting peptidoglycan by the formation of two bonds: (i) a transglycosylase splits the pyrophosphate bond between the undecaprenyl group and the MurNAc of a nascent glycan strand and forms a glycosidic bond between MurNAc and the hydroxyl group of the GlcNAc of the precursor molecule, and (ii) a transpeptidase forms a dd-peptide bond between the carboxyl group of the d-Ala of the precursor and the amino group of an m-Dpm residue present in a peptide moiety of the cell wall. An example of each type of bond attacked by different specific murein hydrolases is also shown. (Adapted from reference 82 with permission.)