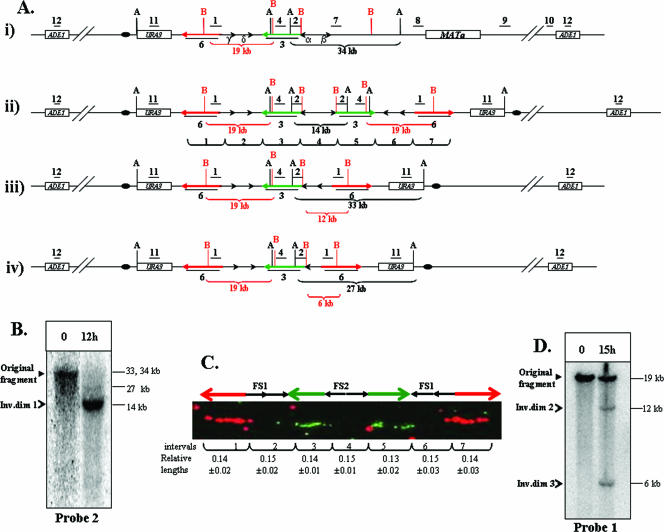
FIG. 3.
Analysis of inverted dicentric dimers formed by interaction between inverted repeats of Ty1. (A) Expected structures of chromosome III of AM919 and repair intermediates derived from this chromosome. (i) The structure of chromosome III of AM919 before the formation of inverted dicentric dimers. Underlined numbers (1, 2, 4, 7, 8, 9, 10, 11, and 12) indicate the positions of probes used to analyze the region (see the supplemental material for a detailed description of all probes). The positions of AvrII (black A) and BspEI (red B) restriction sites are shown; only those sites relevant to the Southern analysis described below are depicted. (ii) The structure of ID1 (370-kb chromosome), resulting from recombination between Ty1α and Ty1β. Probe 3 (shown as a green arrow) is a 14-kb-long probe specific to the region between FS1 and FS2. Probe 6 (shown as a red arrow) is a 14.3-kb-long probe specific to the region centromere proximal to FS1. (iii) The structure of ID2, resulting from recombination between Ty1α and Ty1δ. (iv) The structure of ID3, resulting from recombination between Ty1α and Ty1γ. (B) Analysis of large inverted dimers (Inv.dim) by gel electrophoresis. DNA was extracted from preinduction (0-h) and postinduction (12-h) samples of AM919. The samples were digested with AvrII, and the resulting fragments were separated by gel electrophoresis and examined by Southern analysis with probe 2. As expected from the maps shown in panel A, the DNA fragment in the uninduced sample was about 34 kb. The expected fragment size from the 370-kb ID1 is 14 kb. The expected fragments for ID2 and ID3 are about 33 and 27 kb, respectively. Since the appearance of the 340-kb intermediates is delayed compared to that of the 370-kb intermediate, only small amounts of the 33- and 27-kb fragments are present 12 h after DSB induction. (C) Analysis of large inverted dimers by DNA combing. DNA was extracted from AM919 cells 12 h after the addition of galactose. This DNA was stretched and hybridized to the fluorescent probe 3 (green arrow) and probe 6 (red arrow). The relative lengths of hybridizing regions and the gaps between them were calculated as a fraction of the length between the termini of the red arrows (96 kb). (D) Southern analysis of ID2 and ID3. DNA was extracted from preinduction (0-h) and postinduction (15-h) samples of AM919. The samples were digested with BspEI, and the resulting fragments were examined by Southern analysis using probe 1. Fragments of the expected sizes were found for the wild-type, ID2, and ID3 chromosomes (19, 12, and 6 kb, respectively).