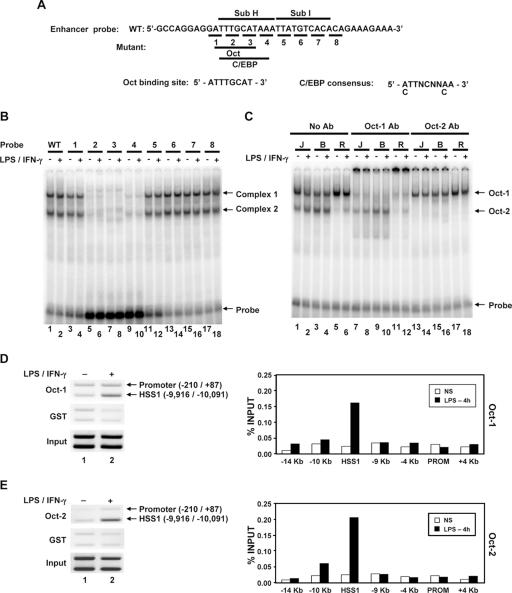
FIG. 6.
Oct-1 and Oct-2 bind the critical Sub H region of the HSS1 enhancer. (A) The DNA sequence of the probe used for gel shift assays is shown. Nucleotides altered in each of the eight mutant probes are underlined. The predicted Oct and C/EBP binding sites within the probe are shown. (B) 32P-labeled wild-type (WT) (lanes 1 and 2) and mutant (lanes 3 to 18) probes (probes 1 to 8) were used for gel shift assays with nuclear extracts from unstimulated B6.129/J2 cells (−) or B6.129/J2 cells stimulated for 4 h with LPS (+). (C) The identities of the two protein-DNA complexes were determined by adding Oct-1 (lanes 7 to 12) and Oct-2 (lanes 13 to 18) antibodies (Ab) to gel shift reaction mixtures. Nuclear extracts were made from J744 (J), B6.129/J2 (B), and RAW264.7 (R) cells. (D) Oct-1 binding to the endogenous HSS1 region was monitored using a ChIP assay. (Left) Formaldehyde-cross-linked chromatin samples prepared from unstimulated peritoneal macrophages (lane 1) or peritoneal macrophages stimulated for 4 h with LPS plus IFN-γ (lane 2) were immunoprecipitated with Oct-1 antibodies (top blot) or control GST antibodies (middle blot). The presence of the HSS1 region and the Il12b promoter in the immunoprecipitates was determined by semiquantitative PCR. Input samples are shown in the bottom blot. (Right) ChIP experiments were performed with bone marrow-derived macrophages, with the Oct-1 antibody immunoprecipitates analyzed by real-time PCR, using a variety of primer pairs that amplify regions spanning the Il12b locus. Similar results were obtained in a second independent experiment (not shown). NS, not stimulated; PROM, promoter. (E) ChIP assays analyzed by both semiquantitative PCR (left) and real-time PCR (right) were performed with Oct-2 or GST antibodies. For both semiquantitative and real-time PCR analysis, chromatin samples were prepared from bone marrow-derived macrophages.