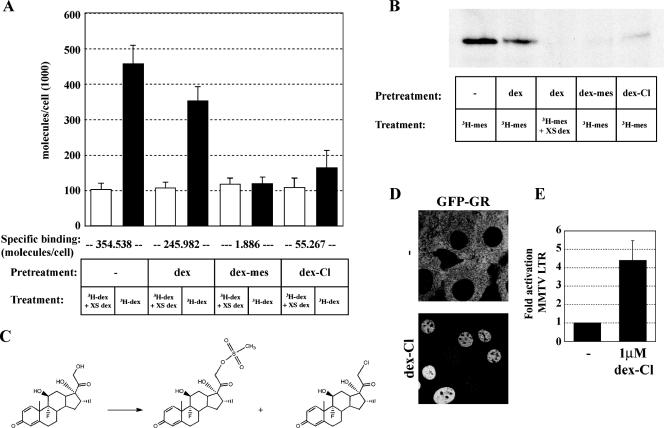
FIG. 5.
Covalent binding of Dex-Mes and Dex-Cl to GR. (A) The number of bound ligand molecules per cell was determined after pretreatment for 45 min with vehicle, 1 μM Dex, 1 μM Dex-Mes, or 1 μM Dex-Cl, followed by three washes for 5 min each with ligand-free medium and subsequent treatment with 100 nM [3H]Dex in the presence or absence of a 100-fold excess of cold Dex. Specific activity and the number of cells per well were used to calculate the number of bound [3H]Dex molecules per cell. Specific binding (below chart) was determined by subtracting nonspecific binding (open bars) in the presence of excess unlabeled Dex from total binding in the presence of 100 nM [3H]Dex (black bars). (B) Fluorograph of covalently labeled GR by [3H]Dex-Mes. Cells were pretreated and washed as described for panel A, followed by a 45-min treatment with 100 nM [3H]Dex-Mes. Protein extracts from whole cells were chromatographed on sodium dodecyl sulfate-polyacrylamide gels and analyzed by fluorography. (C) Synthesis of Dex-Cl (right) and Dex-Mes (middle) from Dex (left). (D) Localization of GFP-GR of untreated cells (top) or cells treated for 45 min with 1 μM Dex-Cl (bottom). (E) Transcriptional activation from the MMTV array is shown as relative mRNA abundance ± SEM for untreated and treated cells as determined by quantitative PCR following a 2-h treatment with 1 μM Dex-Cl.