FIG. 6.
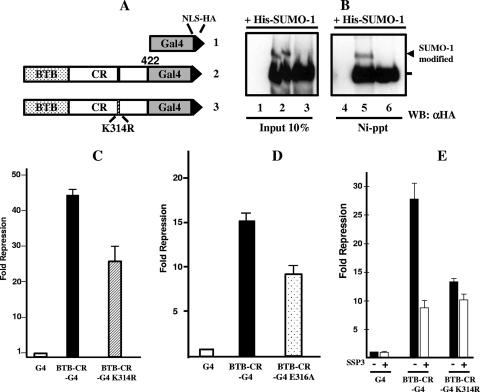
SUMO-1 modification modulates HIC1-mediated transcriptional repression. (A) Schematic structure of the two HIC1 BTB-CR-Gal4 chimeras. Numbers refer to human HIC1 residues. The BTB/POZ domain, the CR, and the Gal4 DBD domain are represented as dotted, white, and gray boxes, respectively. The SUMOylation consensus site is shown as a black line, and the mutated SUMOylation consensus site is shown as a hatched line. A nuclear localization signal (NLS) and an HA epitope tagged at the C-terminal part of the chimeras are shown as black triangles. (B) The BTB-CR-G4 chimera, but not the mutated BTB-CR-G4 K314R chimera, is SUMOylated in vivo. Whole-cell extracts prepared from Cos-7 cells transfected with vectors expressing His-tagged SUMO-1, and the different Gal4 chimeras were either immunoblotted with anti-HA antibody directly (left panel, lanes 1 to 3) or subjected to Ni affinity chromatography prior to Western blot (WB) analysis (right panel, lanes 4 to 6). The arrowhead indicates the position of the SUMO-1-modified BTB-CR-G4 protein. (C) The mutation of K314 in the SUMOylation site reduces the repression capacity of the BTB-CR-Gal4 chimeras. RK13 cells were transiently transfected in triplicate with 200 ng of the indicated Gal4 chimeras and 250 ng of the pG5-luc reporter (schematically drawn in panel A). The luciferase activity was normalized to the β-galactosidase activity of a cotransfected β-OS-lacZ construct (50 ng). After normalization, the data were expressed as Luc activity relative to the activity of pG5-luc with Gal4-NLS-HA expression vector (G4), which was given an arbitrary value of 1. The results are the mean values and standard deviations (error bars) from one independent transfection performed in triplicate that is representative of three independent experiments. (D) The mutation of the consensus E316 residue in the SUMOylation site also reduces the repression capacity of the BTB-CR-Gal4 chimera. A similar experiment was conducted as described above but with expression vectors for the wt BTB-CR-Gal4 chimera or the E316A point mutant. Error bars indicate standard deviations. (E) Expression of the deSUMOylase SENP2/SSP3 impairs the repression potential of the wt and non-SUMOylable K314R BTB-CR-Gal4 chimeras. RK13 cells were transiently transfected in triplicate with 150 ng of the indicated Gal4 chimeras and 200 ng of the pG5-luc reporter alone (−) or in the presence (+) of 200 ng of the SSP3 expression vector. The luciferase activity was normalized to the β-galactosidase activity of a cotransfected β-OS-lacZ construct (50 ng). After normalization, the data were expressed as Luc activity relative to the activity of pG5-luc with the wt chimera in the absence of the SSP3 expression vector, which was given an arbitrary value of 100%. The results are the mean values and standard deviations (error bars) from two independent transfections performed in triplicate.