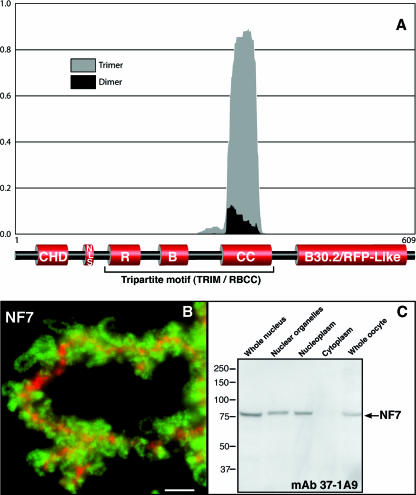
FIG. 1.
NF7 is predicted to form trimers and associates with RNAPII active transcriptional units. (A) The Multicoil algorithm was used on Xenopus NF7 with a scanning window of 28 amino acid residues. A trimeric conformation was strongly predicted, with more than 80% probability for most of the coiled-coil region. This prediction is consistent with the data presented in Fig. 2 and 3, in which NF7 is demonstrated to form homotrimers. (B) Fluorescence micrograph showing a narrow region of an LBC labeled with anti-NF7 MAb 37-1A9 (green) and anti-XCAP-D2 (red). XCAP-D2, which is a subunit of condensin I, is restricted to the transcriptionally inactive heterochromatin domains of the LBCs (3) and was used here to define the chromosomal axes. In contrast, NF7 associates specifically with the RNP matrix of the chromosomal loops, which correspond to RNAPII active transcriptional units. In addition, NF7 is present on nucleoplasmic granules, which are out of focus here because of their small size (0.2 to 0.3 μm). Scale bar, 5 μm. (C) Western blot analysis with MAb 37-1A9 indicates that NF7 is primarily nuclear, and while it is found associated with chromosomes, it is also found soluble in the nucleoplasm. Proteins from 10 whole nuclei and organelles and nucleoplasm from 10 nuclei, five cytoplasms, or five whole oocytes were used. The values on the left are molecular sizes in kilodaltons.