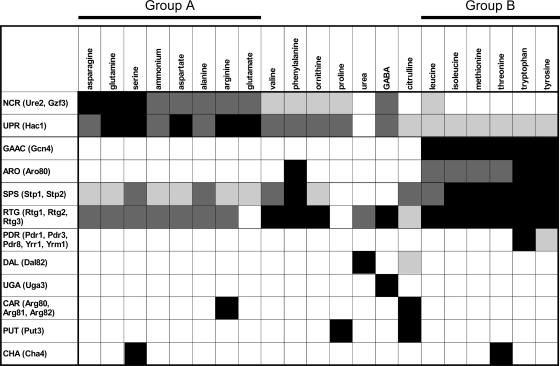
FIG. 9.
Influence of the nitrogen medium on the status of nitrogen-sensitive transcriptional controls. The average levels of expression of the target genes of each indicated regulon were calculated and used to evaluate the degree of activation of the corresponding transcriptional controls (see the text and previous figures). For simplicity's sake, we have defined four degrees of activation: highly active (black squares), moderately active (dark-gray squares), slightly active (light-gray squares), and inactive (white squares). Nitrogen sources are sorted according to the classification presented in Fig. 2A. The activation of transcriptional regulation generally leads to up-regulation of the corresponding target gene. NCR, however, is an exception; in the presence of a good nitrogen source, NCR is active, and this results in the lack of activation of the genes under the positive control of the Gln3 and Gat1/Nil1 transcription factors. ARO, specific induction of genes involved in aromatic amino acid catabolism; SPS, response to extracellular amino acids via the SPS system (Ssy1-Ptr3-Ssy5); PDR, response to drugs; DAL, specific induction of genes involved in allantoin, allantoate, and urea catabolism; UGA, specific induction of genes involved in GABA catabolism; CAR, specific induction of genes involved in arginine catabolism; PUT, specific induction of genes involved in proline catabolism; CHA, specific induction of genes involved in serine and threonine catabolism.