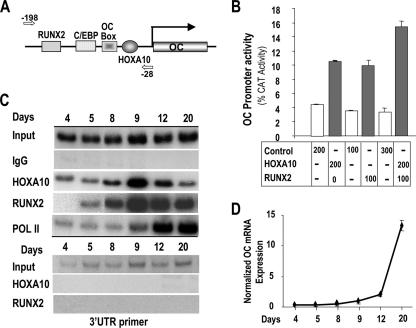
FIG. 7.
HOXA10 is recruited to the OC promoter to directly regulate gene expression. (A) Activation of OC gene transcription by HOXA10. Schematic representation of rat proximal OC promoter segment illustrating the location of the only Hoxa10 consensus motif in the −208-bp 5′ OC fragment. Arrows are the forward (−198) and reverse (−28) OC primers used in ChIP analysis. (B) The mouse preosteoblast cell line MC3T3-E1 was cotransfected with either empty vector (control) or Hoxa10 and Runx2 expression plasmids as indicated and with the proximal OC promoter (−208 OC-CAT) by use of FuGENE6 (Roche Molecular Biologicals). Control cells contained the indicated amount of backbone plasmid (pGL3). Cells were harvested 24 h posttransfection for CAT assay. Promoter activity was calculated from values equivalent to those for six samples and expressed as percent CAT activity. (C) In vivo occupancy of HOXA10 on the OC gene. Cross-linked chromatin samples from primary rat calvarium-derived osteoblast cultures at the indicated stages of differentiation (days 4, 5, 8, 9, 12, and 20) were used for immunoprecipitation reactions with 2 μg of HOXA10, RUNX2, Pol II, and nonspecific antibody (IgG). The pull-down DNA fragments were purified and assayed by PCR. Input represents 1% of each chromatin fraction used for immunoprecipitation. The ChIP data presented are representative of multiple experiments in which all the time point data were derived from the same osteoblast preparation. The lower panel shows the control for ChIP primer specificity. (D) Expression of OC mRNA (QPCR) during differentiation of primary calvarial osteoblasts at the indicated days. C/EBP, CCAAT/enhancer-binding proteins.