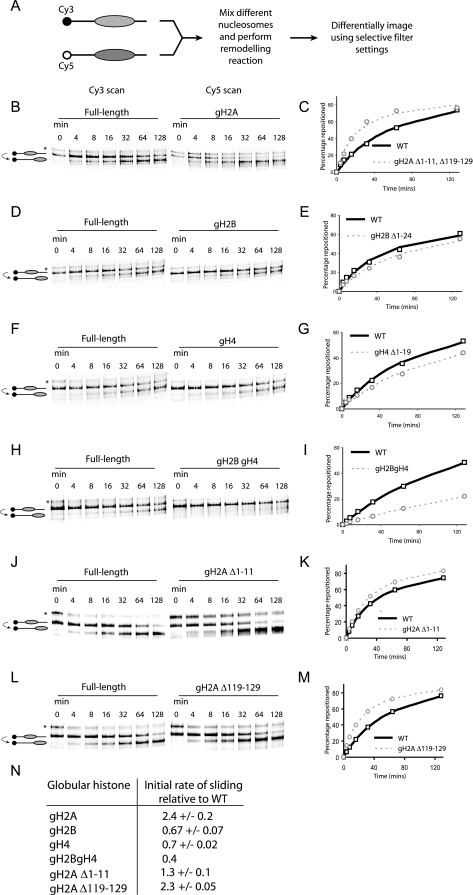
FIG. 1.
Effect of histone tail deletions on thermal nucleosome sliding. (A) Two picomoles of truncated and full-length octamers was assembled onto differentially labeled fluorescent DNA, mixed, and incubated at 47°C for the specified amount of time. Nucleosomes at different translational positions were separated on a native polyacrylamide gel, and the individual types of nucleosomes were visualized using the appropriate excitation and emission settings as illustrated. The majority of nucleosomes assembled at a position +70 relative to the MMTV transcriptional start site; following thermal incubation these equilibrated to a more favorable location at +22 (16). However, a proportion of nucleosomes (indicated by an asterisk) was deposited at a less strongly favored location at +47 (53). (B to M) Individual tail deletions have distinct effects on the rate of nucleosome repositioning. The graphs show the fraction of nucleosomes repositioned as a function of time. The data points are the average of three independent repeats, with the exception of panel F, and the curve describes the line of best fit used to calculate the initial rates. (N) Initial rate of repositioning for globular nucleosomes relative to a full-length control. WT, wild type.