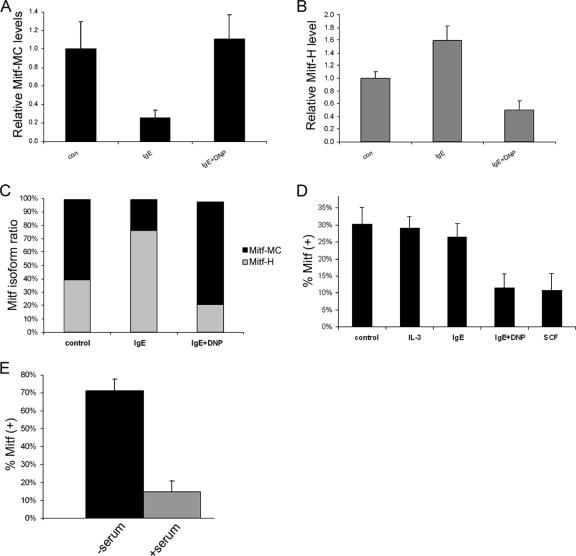
FIG. 2.
Influence of physiological stimuli on Mitf isoform expression. (A and B) Real-time PCR analysis of Mitf-MC (A) and Mitf-H (B) expression in BMMC. Cells were activated with either IgE alone or IgE followed by DNP (IgE + DNP). con, control. Results represent mean ± standard error (n = 3). (C) Results of real-time PCR analysis of Mitf-H and Mitf-MC isoforms showing the relative amounts of the two isoforms expressed as a percentage of the total Mitf. (D) Analysis of the expression of exon 6a in BMMC. Cells were activated with either IL-3, IgE alone, IgE followed by DNP (IgE + DNP), or SCF. Exon 5 to exon 7 were amplified by PCR (25 cycles) and separated on 8% acrylamide gel, and the ratio between the positive and negative isoforms was determined by densitometry. Results are expressed as percentage of Mitf-(+) out of total Mitf. Results represent the mean ± standard error (n = 3). (E) Analysis of the expression of exon 6a in H9C2 cardiomyoctes. Cells were grown initially in Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal calf serum (FCS). The medium was then replaced with either serum-free DMEM or fresh DMEM with FCS for 6 h. Results are expressed as percentage of Mitf-(+) out of total Mitf. Results represent the mean ± standard error (n = 5).