FIG. 2.
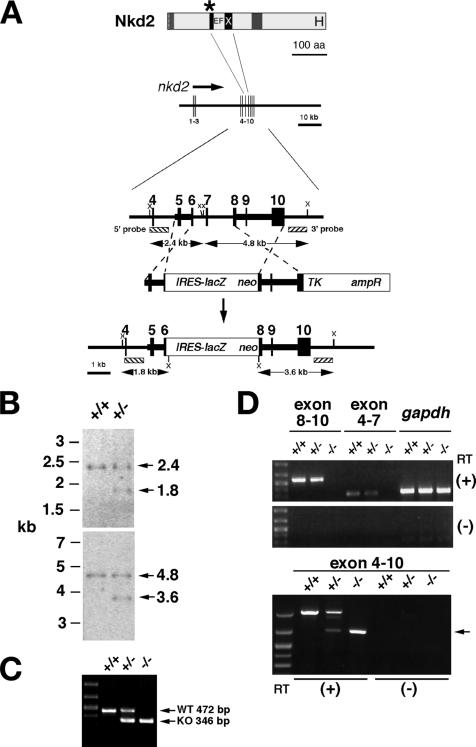
Targeting of mouse nkd2. (A) Schematic of Nkd2 protein above the nkd2 genomic region as described for Fig. 1. Homologous recombination replaced exons 6 to 8 with the IRES-lacZ/neo cassette. Enzyme site: X, XbaI. aa, amino acids. (B) Southern blotting of tail DNAs from mice of the indicated genotypes cut with XbaI (top and bottom) and probed with 5′ (top) or 3′ (bottom) external probes as designated in panel A. (C) PCR genotyping with tail DNA from mice of the indicated genotypes amplified by primers specific for the wild-type (WT) and nkd2lacZ (KO) chromosomes. (D) RT-PCR analysis of brain RNA from adult mice of the indicated nkd2 genotypes with primers spanning the designated nkd2 exons or the control gene gapdh, with (+) or without (−) reverse transcriptase (RT). Markers are loaded in the leftmost lanes. (Top) −/− reactions lack specific bands spanning exons 8 to 10 (579 bp) or 4 to 7 (323 bp) that were deleted by the targeting event. gapdh (350 bp) is a positive control that is amplified from all three samples. (Bottom) +/+ mice have a 1,025-bp band corresponding to the full-length exon 4 to 10 product. +/− mice have 1,025- and 699-bp bands, while −/− mice only have the 699-bp band (arrow). Sequencing of the 699-bp band revealed exon 5-9-10 splicing, predicting the synthesis of an out-of-frame, truncated protein (data not shown).