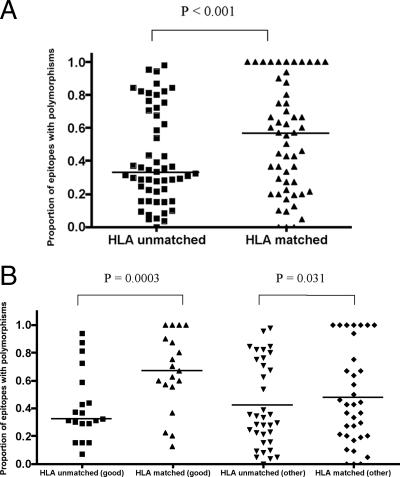
FIG. 1.
Proportion of optimal epitopes containing polymorphisms in HLA-matched and HLA-unmatched (control) patients. (A) For each of the 54 HLA class I-restricted optimal epitopes in the Swiss cohort, the proportion of variant sequences was calculated in patients expressing the relevant HLA class I allele (matched) and in a control group of patients not carrying that allele (unmatched). The figure shows the distribution of variation in the matched and unmatched groups (Wilcoxon signed-rank test). Statistical significance remained after removal of epitopes with 100% variation from the analysis (P = 0.0007) (data not shown). (B) Epitopes were divided according to whether they were restricted by good (HLA-A*11, -B*27, -B*51, -B*57, and -B*58, which are associated with clinical advantage) or other (not associated with clinical advantage) HLA class I alleles. The proportion of variants in each group was compared using the Wilcoxon signed-rank test.