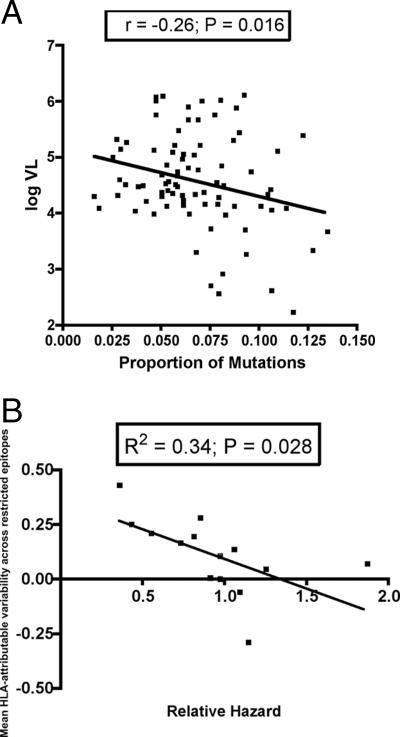
FIG. 5.
Epitope variability predicts plasma viral load and relative hazard of disease progression. (A) For each patient the proportion of sequence variants in 54 optimal epitopes in the Gag, Pol, and Nef genes was calculated and correlated with baseline plasma viral load, using Pearson's correlation coefficient. (B) For each epitope the difference in the proportion of variant epitopes in HLA-matched and -unmatched patients was calculated. A mean value for each HLA class I molecule was then calculated from all the epitopes restricted by each HLA class I allele. These values were used as a measure of polymorphic variability associated with each HLA class I molecule and correlated negatively with the associated relative hazard of disease progression for each HLA class I allele.