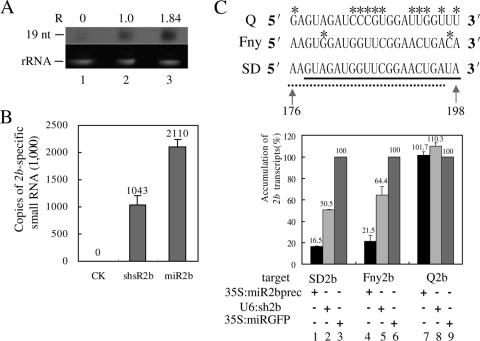
FIG. 2.
Artificial 2b-specific small RNAs down-regulated the expression of the 2b gene with sequence specificity. (A) Solution hybridization analysis of artificial sRNAs generated in agroinfiltrated N. benthamiana leaves. sRNAs hybridized with a 32P-labeled miR2b- and shsR2b-specific probe and protected from RNase A/T1 digestion were resolved by 15% polyacrylamide gel electrophoresis and detected by autoradiography (top). The migration position of a 19-nt RNA oligonucleotide is indicated on the left. The relative intensity (R) of the hybridizing bands determined by density scanning is shown above the panel. Ethidium bromide-stained 5.8S rRNA, indicating an equal amount of RNA (1 μg), was used in solution hybridization (bottom). Lane 1, control strain-infiltrated leaves; lane 2, U6:sh2b-infiltrated leaves; lane 3, 35S:miR2bprec-infiltrated leaves. (B) Real-time RT-PCR quantification of the expression levels of artificial sRNAs. Total RNA extracted from the agroinfiltrated area of N. benthamiana leaves was used as the template in the RT reaction. The arbitrary copy number of 2b-specific sRNAs is presented relative to 1 × 106 copies of the EF1α transcript. Error bars represent standard deviations of three determinations, and similar results were obtained in two independent RT-PCR experiments. CK, control strain-infiltrated leaves; shsR2b, U6:sh2b-infiltrated leaves; miR2b, 35S:miR2bprec-infiltrated leaves. (C) Artificial sRNAs inhibited the expression of the 2b genes with sequence specificity. The target regions in 2b genes of three CMV strains (Q, Fny, and SD) are shown (top). The nucleotides unpaired with the sequence of miR2b (thick line) or shsR2b (broken line) are marked with asterisks. The accumulation levels of the 35S promoter-produced 2b transcripts (SD 2b, Fny 2b, and Q 2b) in N. benthamiana leaves coinfiltrated with the miR2b-producing (35S:miR2bprec) or the shsR2b-producing (U6:sh2b) Agrobacterium strain were determined by real-time RT-PCR and compared to levels when a miRGFP-producing strain (35S:miRGFP) was coinfiltrated (bottom). Error bars represent standard deviations of three determinations, and similar results were obtained in three independent RT-PCR experiments.