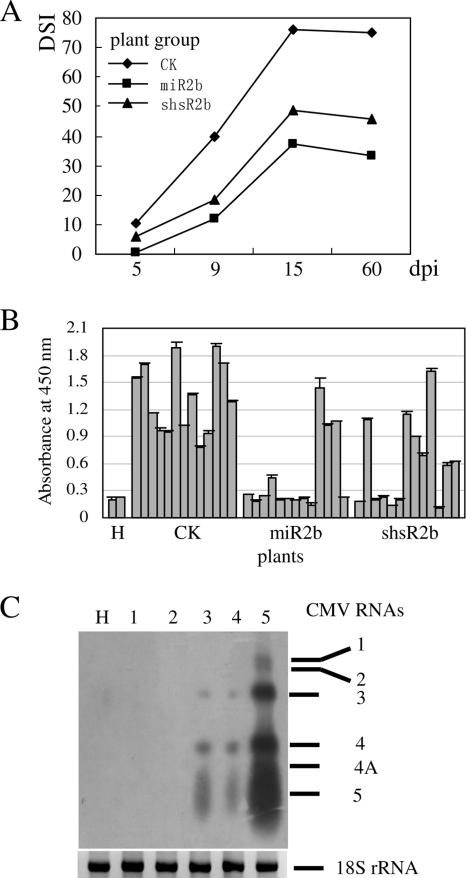
FIG. 4.
Transgenic plants expressing miR2b or shsR2b exhibited resistance to CMV infection. (A) Symptom development in T1 progeny of 16 p35S-miR2bprec-, 14 pU6-sh2b-, and 18 control vector-transformed plant lines after CMV inoculation. Three to five T1 plants, each from independent T0 lines, were used in the challenge experiments. DSIs of each group of transgenic plants (abbreviated as miR2b, shsR2b and CK, respectively) were calculated based on the disease index grade of all individual plants within each group scored at 5, 9, 15, and 60 dpi. (B) Accumulation of CMV CP in CMV-inoculated transgenic plants. From each transgenic group, 13 representative T1 plants, each from an individual T0 line, were selected at 9 dpi, and the level of CMV CP in upper uninoculated leaves of each plant was determined by ELISA. Two uninoculated tobacco plants (H) were used as a blank control in the ELISA. Error bars represent the standard deviations of two repeat assays. (C) Northern blot analysis of CMV RNAs accumulated in CMV-inoculated transgenic plants. Total RNA from upper uninoculated leaves at 15 dpi was used in the experiments. Lanes 1 and 2, an miR2b-plant (m-1-2) and an shsR2b-plant (s-4-13), respectively, showing no symptoms; lanes 3 and 4, an miR2b-plant (m-19-33) and a shsR2b-plant (s-25-3), respectively, showing mild symptoms; lane 5, control vector-transformed plant; lane H, uninoculated tobacco plant. On the right, the migration positions of CMV RNAs 1, 2, 3, 4, 4A, and 5 are indicated. Ethidium bromide-stained 18S rRNA was used as an RNA loading control (bottom).